Mengyang Zhao
Logic-Guided Multistage Inference for Explainable Multidefendant Judgment Prediction
Jan 19, 2026Abstract:Crime disrupts societal stability, making law essential for balance. In multidefendant cases, assigning responsibility is complex and challenges fairness, requiring precise role differentiation. However, judicial phrasing often obscures the roles of the defendants, hindering effective AI-driven analyses. To address this issue, we incorporate sentencing logic into a pretrained Transformer encoder framework to enhance the intelligent assistance in multidefendant cases while ensuring legal interpretability. Within this framework an oriented masking mechanism clarifies roles and a comparative data construction strategy improves the model's sensitivity to culpability distinctions between principals and accomplices. Predicted guilt labels are further incorporated into a regression model through broadcasting, consolidating crime descriptions and court views. Our proposed masked multistage inference (MMSI) framework, evaluated on the custom IMLJP dataset for intentional injury cases, achieves significant accuracy improvements, outperforming baselines in role-based culpability differentiation. This work offers a robust solution for enhancing intelligent judicial systems, with publicly code available.
Human + AI for Accelerating Ad Localization Evaluation
Sep 16, 2025Abstract:Adapting advertisements for multilingual audiences requires more than simple text translation; it demands preservation of visual consistency, spatial alignment, and stylistic integrity across diverse languages and formats. We introduce a structured framework that combines automated components with human oversight to address the complexities of advertisement localization. To the best of our knowledge, this is the first work to integrate scene text detection, inpainting, machine translation (MT), and text reimposition specifically for accelerating ad localization evaluation workflows. Qualitative results across six locales demonstrate that our approach produces semantically accurate and visually coherent localized advertisements, suitable for deployment in real-world workflows.
Interpretable Oracle Bone Script Decipherment through Radical and Pictographic Analysis with LVLMs
Aug 13, 2025Abstract:As the oldest mature writing system, Oracle Bone Script (OBS) has long posed significant challenges for archaeological decipherment due to its rarity, abstractness, and pictographic diversity. Current deep learning-based methods have made exciting progress on the OBS decipherment task, but existing approaches often ignore the intricate connections between glyphs and the semantics of OBS. This results in limited generalization and interpretability, especially when addressing zero-shot settings and undeciphered OBS. To this end, we propose an interpretable OBS decipherment method based on Large Vision-Language Models, which synergistically combines radical analysis and pictograph-semantic understanding to bridge the gap between glyphs and meanings of OBS. Specifically, we propose a progressive training strategy that guides the model from radical recognition and analysis to pictographic analysis and mutual analysis, thus enabling reasoning from glyph to meaning. We also design a Radical-Pictographic Dual Matching mechanism informed by the analysis results, significantly enhancing the model's zero-shot decipherment performance. To facilitate model training, we propose the Pictographic Decipherment OBS Dataset, which comprises 47,157 Chinese characters annotated with OBS images and pictographic analysis texts. Experimental results on public benchmarks demonstrate that our approach achieves state-of-the-art Top-10 accuracy and superior zero-shot decipherment capabilities. More importantly, our model delivers logical analysis processes, possibly providing archaeologically valuable reference results for undeciphered OBS, and thus has potential applications in digital humanities and historical research. The dataset and code will be released in https://github.com/PKXX1943/PD-OBS.
From Intent to Execution: Multimodal Chain-of-Thought Reinforcement Learning for Precise CAD Code Generation
Aug 13, 2025Abstract:Computer-Aided Design (CAD) plays a vital role in engineering and manufacturing, yet current CAD workflows require extensive domain expertise and manual modeling effort. Recent advances in large language models (LLMs) have made it possible to generate code from natural language, opening new opportunities for automating parametric 3D modeling. However, directly translating human design intent into executable CAD code remains highly challenging, due to the need for logical reasoning, syntactic correctness, and numerical precision. In this work, we propose CAD-RL, a multimodal Chain-of-Thought (CoT) guided reinforcement learning post training framework for CAD modeling code generation. Our method combines CoT-based Cold Start with goal-driven reinforcement learning post training using three task-specific rewards: executability reward, geometric accuracy reward, and external evaluation reward. To ensure stable policy learning under sparse and high-variance reward conditions, we introduce three targeted optimization strategies: Trust Region Stretch for improved exploration, Precision Token Loss for enhanced dimensions parameter accuracy, and Overlong Filtering to reduce noisy supervision. To support training and benchmarking, we release ExeCAD, a noval dataset comprising 16,540 real-world CAD examples with paired natural language and structured design language descriptions, executable CADQuery scripts, and rendered 3D models. Experiments demonstrate that CAD-RL achieves significant improvements in reasoning quality, output precision, and code executability over existing VLMs.
CrowdTrack: A Benchmark for Difficult Multiple Pedestrian Tracking in Real Scenarios
Jul 03, 2025Abstract:Multi-object tracking is a classic field in computer vision. Among them, pedestrian tracking has extremely high application value and has become the most popular research category. Existing methods mainly use motion or appearance information for tracking, which is often difficult in complex scenarios. For the motion information, mutual occlusions between objects often prevent updating of the motion state; for the appearance information, non-robust results are often obtained due to reasons such as only partial visibility of the object or blurred images. Although learning how to perform tracking in these situations from the annotated data is the simplest solution, the existing MOT dataset fails to satisfy this solution. Existing methods mainly have two drawbacks: relatively simple scene composition and non-realistic scenarios. Although some of the video sequences in existing dataset do not have the above-mentioned drawbacks, the number is far from adequate for research purposes. To this end, we propose a difficult large-scale dataset for multi-pedestrian tracking, shot mainly from the first-person view and all from real-life complex scenarios. We name it ``CrowdTrack'' because there are numerous objects in most of the sequences. Our dataset consists of 33 videos, containing a total of 5,185 trajectories. Each object is annotated with a complete bounding box and a unique object ID. The dataset will provide a platform to facilitate the development of algorithms that remain effective in complex situations. We analyzed the dataset comprehensively and tested multiple SOTA models on our dataset. Besides, we analyzed the performance of the foundation models on our dataset. The dataset and project code is released at: https://github.com/loseevaya/CrowdTrack .
ChatReID: Open-ended Interactive Person Retrieval via Hierarchical Progressive Tuning for Vision Language Models
Feb 27, 2025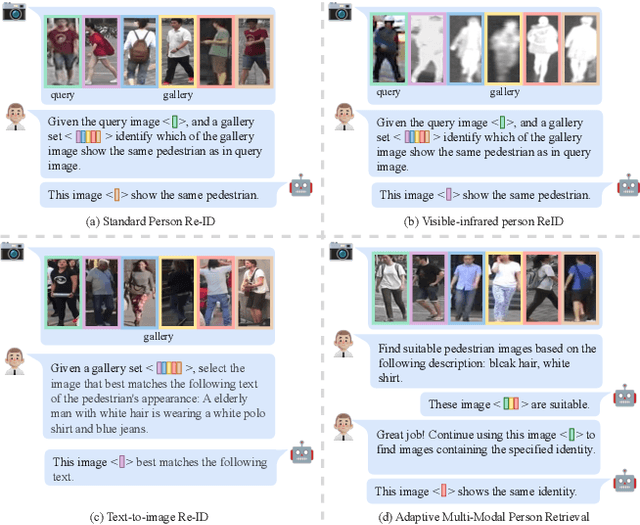
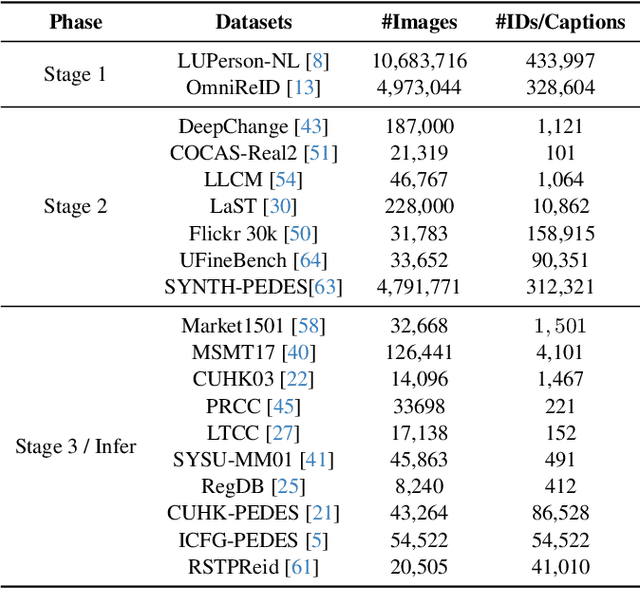
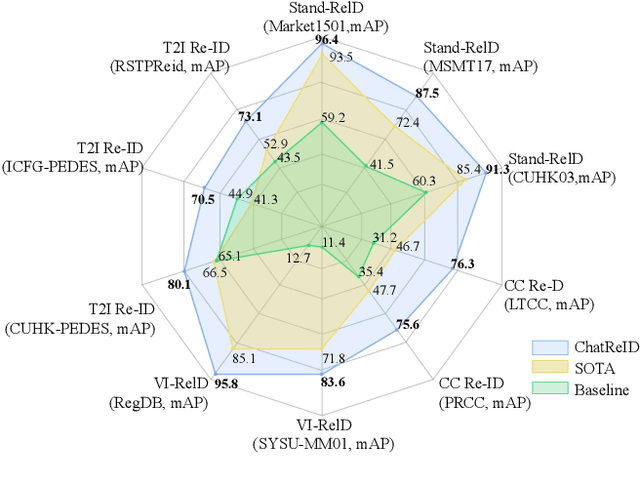
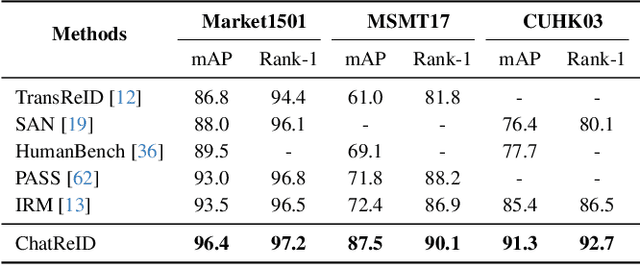
Abstract:Person re-identification (Re-ID) is a critical task in human-centric intelligent systems, enabling consistent identification of individuals across different camera views using multi-modal query information. Recent studies have successfully integrated LVLMs with person Re-ID, yielding promising results. However, existing LVLM-based methods face several limitations. They rely on extracting textual embeddings from fixed templates, which are used either as intermediate features for image representation or for prompt tuning in domain-specific tasks. Furthermore, they are unable to adopt the VQA inference format, significantly restricting their broader applicability. In this paper, we propose a novel, versatile, one-for-all person Re-ID framework, ChatReID. Our approach introduces a Hierarchical Progressive Tuning (HPT) strategy, which ensures fine-grained identity-level retrieval by progressively refining the model's ability to distinguish pedestrian identities. Extensive experiments demonstrate that our approach outperforms SOTA methods across ten benchmarks in four different Re-ID settings, offering enhanced flexibility and user-friendliness. ChatReID provides a scalable, practical solution for real-world person Re-ID applications, enabling effective multi-modal interaction and fine-grained identity discrimination.
STNMamba: Mamba-based Spatial-Temporal Normality Learning for Video Anomaly Detection
Dec 28, 2024Abstract:Video anomaly detection (VAD) has been extensively researched due to its potential for intelligent video systems. However, most existing methods based on CNNs and transformers still suffer from substantial computational burdens and have room for improvement in learning spatial-temporal normality. Recently, Mamba has shown great potential for modeling long-range dependencies with linear complexity, providing an effective solution to the above dilemma. To this end, we propose a lightweight and effective Mamba-based network named STNMamba, which incorporates carefully designed Mamba modules to enhance the learning of spatial-temporal normality. Firstly, we develop a dual-encoder architecture, where the spatial encoder equipped with Multi-Scale Vision Space State Blocks (MS-VSSB) extracts multi-scale appearance features, and the temporal encoder employs Channel-Aware Vision Space State Blocks (CA-VSSB) to capture significant motion patterns. Secondly, a Spatial-Temporal Interaction Module (STIM) is introduced to integrate spatial and temporal information across multiple levels, enabling effective modeling of intrinsic spatial-temporal consistency. Within this module, the Spatial-Temporal Fusion Block (STFB) is proposed to fuse the spatial and temporal features into a unified feature space, and the memory bank is utilized to store spatial-temporal prototypes of normal patterns, restricting the model's ability to represent anomalies. Extensive experiments on three benchmark datasets demonstrate that our STNMamba achieves competitive performance with fewer parameters and lower computational costs than existing methods.
LGN-Net: Local-Global Normality Network for Video Anomaly Detection
Nov 21, 2022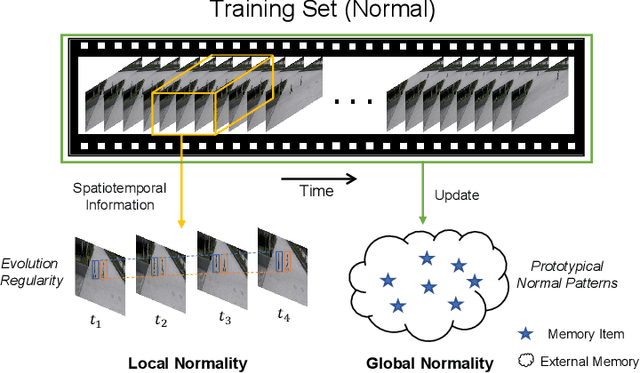
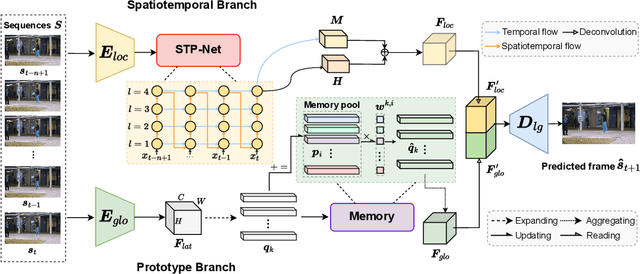
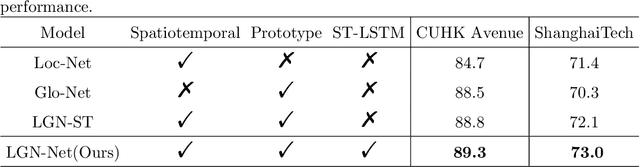
Abstract:Video anomaly detection (VAD) has been intensively studied for years because of its potential applications in intelligent video systems. Existing unsupervised VAD methods tend to learn normality from training sets consisting of only normal videos and regard instances deviating from such normality as anomalies. However, they often consider only local or global normality. Some of them focus on learning local spatiotemporal representations from consecutive frames in video clips to enhance the representation for normal events. But powerful representation allows these methods to represent some anomalies and causes missed detections. In contrast, the other methods are devoted to memorizing global prototypical patterns of whole training videos to weaken the generalization for anomalies, which also restricts them to represent diverse normal patterns and causes false alarms. To this end, we propose a two-branch model, Local-Global Normality Network (LGN-Net), to learn local and global normality simultaneously. Specifically, one branch learns the evolution regularities of appearance and motion from consecutive frames as local normality utilizing a spatiotemporal prediction network, while the other branch memorizes prototype features of the whole videos as global normality by a memory module. LGN-Net achieves a balance of representing normal and abnormal instances by fusing local and global normality. The fused normality enables our model more generalized to various scenes compared to exploiting single normality. Experiments demonstrate the effectiveness and superior performance of our method. The code is available online: https://github.com/Myzhao1999/LGN-Net.
Exploiting Spatial-temporal Correlations for Video Anomaly Detection
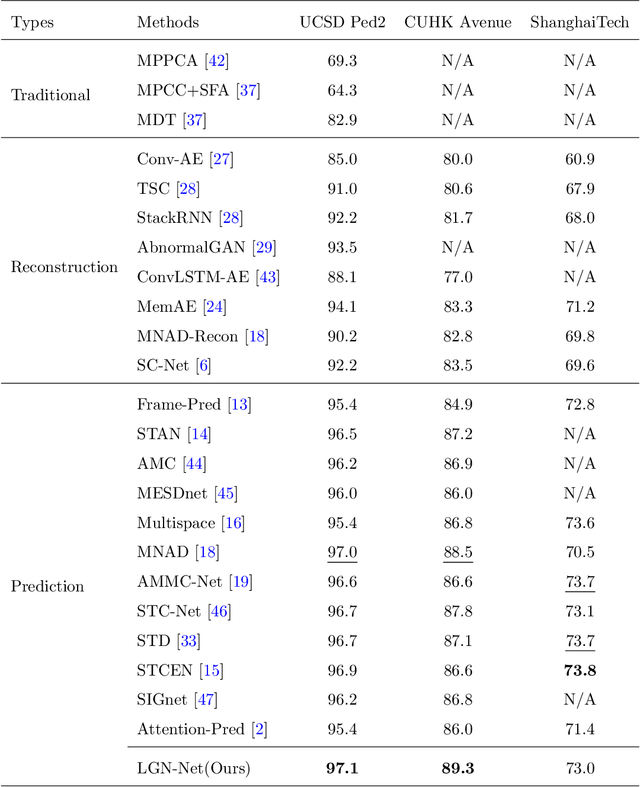
Nov 02, 2022Abstract:Video anomaly detection (VAD) remains a challenging task in the pattern recognition community due to the ambiguity and diversity of abnormal events. Existing deep learning-based VAD methods usually leverage proxy tasks to learn the normal patterns and discriminate the instances that deviate from such patterns as abnormal. However, most of them do not take full advantage of spatial-temporal correlations among video frames, which is critical for understanding normal patterns. In this paper, we address unsupervised VAD by learning the evolution regularity of appearance and motion in the long and short-term and exploit the spatial-temporal correlations among consecutive frames in normal videos more adequately. Specifically, we proposed to utilize the spatiotemporal long short-term memory (ST-LSTM) to extract and memorize spatial appearances and temporal variations in a unified memory cell. In addition, inspired by the generative adversarial network, we introduce a discriminator to perform adversarial learning with the ST-LSTM to enhance the learning capability. Experimental results on standard benchmarks demonstrate the effectiveness of spatial-temporal correlations for unsupervised VAD. Our method achieves competitive performance compared to the state-of-the-art methods with AUCs of 96.7%, 87.8%, and 73.1% on the UCSD Ped2, CUHK Avenue, and ShanghaiTech, respectively.
Compound Figure Separation of Biomedical Images: Mining Large Datasets for Self-supervised Learning
Aug 30, 2022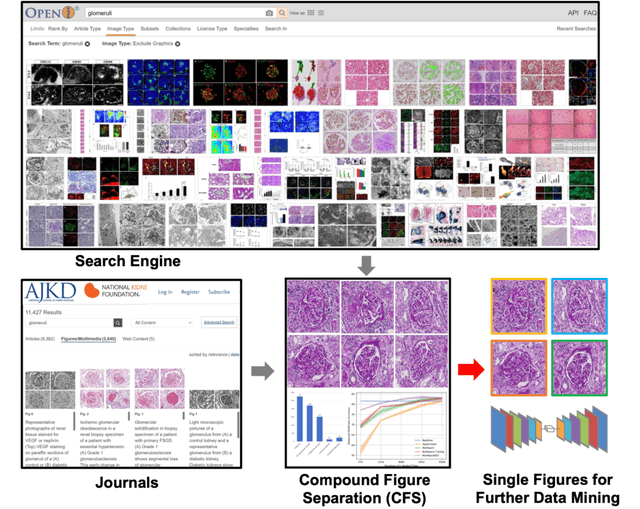
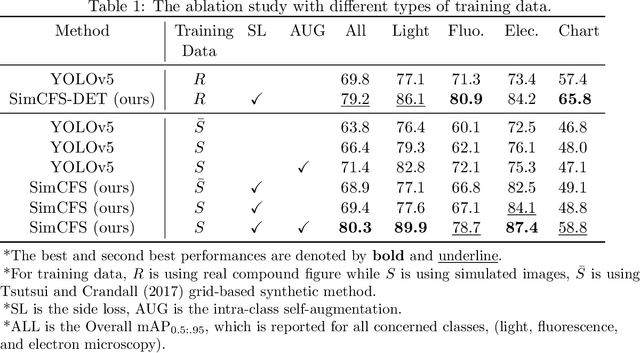
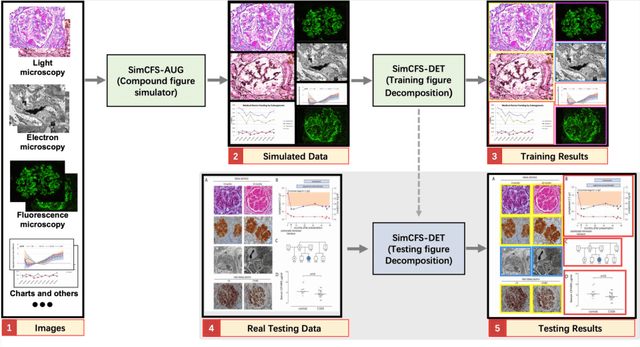
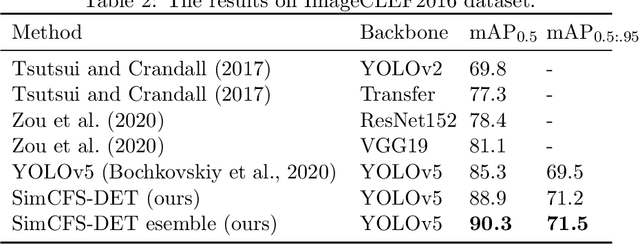
Abstract:With the rapid development of self-supervised learning (e.g., contrastive learning), the importance of having large-scale images (even without annotations) for training a more generalizable AI model has been widely recognized in medical image analysis. However, collecting large-scale task-specific unannotated data at scale can be challenging for individual labs. Existing online resources, such as digital books, publications, and search engines, provide a new resource for obtaining large-scale images. However, published images in healthcare (e.g., radiology and pathology) consist of a considerable amount of compound figures with subplots. In order to extract and separate compound figures into usable individual images for downstream learning, we propose a simple compound figure separation (SimCFS) framework without using the traditionally required detection bounding box annotations, with a new loss function and a hard case simulation. Our technical contribution is four-fold: (1) we introduce a simulation-based training framework that minimizes the need for resource extensive bounding box annotations; (2) we propose a new side loss that is optimized for compound figure separation; (3) we propose an intra-class image augmentation method to simulate hard cases; and (4) to the best of our knowledge, this is the first study that evaluates the efficacy of leveraging self-supervised learning with compound image separation. From the results, the proposed SimCFS achieved state-of-the-art performance on the ImageCLEF 2016 Compound Figure Separation Database. The pretrained self-supervised learning model using large-scale mined figures improved the accuracy of downstream image classification tasks with a contrastive learning algorithm. The source code of SimCFS is made publicly available at https://github.com/hrlblab/ImageSeperation.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://www.melba-journal.org/papers/2022:025.html. arXiv admin note: substantial text overlap with arXiv:2107.08650
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge