Zuhayr Asad
Digital Modeling on Large Kernel Metamaterial Neural Network
Jul 21, 2023Abstract:Deep neural networks (DNNs) utilized recently are physically deployed with computational units (e.g., CPUs and GPUs). Such a design might lead to a heavy computational burden, significant latency, and intensive power consumption, which are critical limitations in applications such as the Internet of Things (IoT), edge computing, and the usage of drones. Recent advances in optical computational units (e.g., metamaterial) have shed light on energy-free and light-speed neural networks. However, the digital design of the metamaterial neural network (MNN) is fundamentally limited by its physical limitations, such as precision, noise, and bandwidth during fabrication. Moreover, the unique advantages of MNN's (e.g., light-speed computation) are not fully explored via standard 3x3 convolution kernels. In this paper, we propose a novel large kernel metamaterial neural network (LMNN) that maximizes the digital capacity of the state-of-the-art (SOTA) MNN with model re-parametrization and network compression, while also considering the optical limitation explicitly. The new digital learning scheme can maximize the learning capacity of MNN while modeling the physical restrictions of meta-optic. With the proposed LMNN, the computation cost of the convolutional front-end can be offloaded into fabricated optical hardware. The experimental results on two publicly available datasets demonstrate that the optimized hybrid design improved classification accuracy while reducing computational latency. The development of the proposed LMNN is a promising step towards the ultimate goal of energy-free and light-speed AI.
Feasibility of Universal Anomaly Detection without Knowing the Abnormality in Medical Images
Jul 03, 2023



Abstract:Many anomaly detection approaches, especially deep learning methods, have been recently developed to identify abnormal image morphology by only employing normal images during training. Unfortunately, many prior anomaly detection methods were optimized for a specific "known" abnormality (e.g., brain tumor, bone fraction, cell types). Moreover, even though only the normal images were used in the training process, the abnormal images were oftenly employed during the validation process (e.g., epoch selection, hyper-parameter tuning), which might leak the supposed ``unknown" abnormality unintentionally. In this study, we investigated these two essential aspects regarding universal anomaly detection in medical images by (1) comparing various anomaly detection methods across four medical datasets, (2) investigating the inevitable but often neglected issues on how to unbiasedly select the optimal anomaly detection model during the validation phase using only normal images, and (3) proposing a simple decision-level ensemble method to leverage the advantage of different kinds of anomaly detection without knowing the abnormality. The results of our experiments indicate that none of the evaluated methods consistently achieved the best performance across all datasets. Our proposed method enhanced the robustness of performance in general (average AUC 0.956).
Robust Fiber ODF Estimation Using Deep Constrained Spherical Deconvolution for Diffusion MRI
Jun 05, 2023Abstract:Diffusion-weighted magnetic resonance imaging (DW-MRI) is a critical imaging method for capturing and modeling tissue microarchitecture at a millimeter scale. A common practice to model the measured DW-MRI signal is via fiber orientation distribution function (fODF). This function is the essential first step for the downstream tractography and connectivity analyses. With recent advantages in data sharing, large-scale multi-site DW-MRI datasets are being made available for multi-site studies. However, measurement variabilities (e.g., inter- and intra-site variability, hardware performance, and sequence design) are inevitable during the acquisition of DW-MRI. Most existing model-based methods (e.g., constrained spherical deconvolution (CSD)) and learning based methods (e.g., deep learning (DL)) do not explicitly consider such variabilities in fODF modeling, which consequently leads to inferior performance on multi-site and/or longitudinal diffusion studies. In this paper, we propose a novel data-driven deep constrained spherical deconvolution method to explicitly constrain the scan-rescan variabilities for a more reproducible and robust estimation of brain microstructure from repeated DW-MRI scans. Specifically, the proposed method introduces a new 3D volumetric scanner-invariant regularization scheme during the fODF estimation. We study the Human Connectome Project (HCP) young adults test-retest group as well as the MASiVar dataset (with inter- and intra-site scan/rescan data). The Baltimore Longitudinal Study of Aging (BLSA) dataset is employed for external validation. From the experimental results, the proposed data-driven framework outperforms the existing benchmarks in repeated fODF estimation. The proposed method is assessing the downstream connectivity analysis and shows increased performance in distinguishing subjects with different biomarkers.
Democratizing Pathological Image Segmentation with Lay Annotators via Molecular-empowered Learning
May 31, 2023Abstract:Multi-class cell segmentation in high-resolution Giga-pixel whole slide images (WSI) is critical for various clinical applications. Training such an AI model typically requires labor-intensive pixel-wise manual annotation from experienced domain experts (e.g., pathologists). Moreover, such annotation is error-prone when differentiating fine-grained cell types (e.g., podocyte and mesangial cells) via the naked human eye. In this study, we assess the feasibility of democratizing pathological AI deployment by only using lay annotators (annotators without medical domain knowledge). The contribution of this paper is threefold: (1) We proposed a molecular-empowered learning scheme for multi-class cell segmentation using partial labels from lay annotators; (2) The proposed method integrated Giga-pixel level molecular-morphology cross-modality registration, molecular-informed annotation, and molecular-oriented segmentation model, so as to achieve significantly superior performance via 3 lay annotators as compared with 2 experienced pathologists; (3) A deep corrective learning (learning with imperfect label) method is proposed to further improve the segmentation performance using partially annotated noisy data. From the experimental results, our learning method achieved F1 = 0.8496 using molecular-informed annotations from lay annotators, which is better than conventional morphology-based annotations (F1 = 0.7051) from experienced pathologists. Our method democratizes the development of a pathological segmentation deep model to the lay annotator level, which consequently scales up the learning process similar to a non-medical computer vision task. The official implementation and cell annotations are publicly available at https://github.com/hrlblab/MolecularEL.
An Accelerated Pipeline for Multi-label Renal Pathology Image Segmentation at the Whole Slide Image Level
May 23, 2023Abstract:Deep-learning techniques have been used widely to alleviate the labour-intensive and time-consuming manual annotation required for pixel-level tissue characterization. Our previous study introduced an efficient single dynamic network - Omni-Seg - that achieved multi-class multi-scale pathological segmentation with less computational complexity. However, the patch-wise segmentation paradigm still applies to Omni-Seg, and the pipeline is time-consuming when providing segmentation for Whole Slide Images (WSIs). In this paper, we propose an enhanced version of the Omni-Seg pipeline in order to reduce the repetitive computing processes and utilize a GPU to accelerate the model's prediction for both better model performance and faster speed. Our proposed method's innovative contribution is two-fold: (1) a Docker is released for an end-to-end slide-wise multi-tissue segmentation for WSIs; and (2) the pipeline is deployed on a GPU to accelerate the prediction, achieving better segmentation quality in less time. The proposed accelerated implementation reduced the average processing time (at the testing stage) on a standard needle biopsy WSI from 2.3 hours to 22 minutes, using 35 WSIs from the Kidney Tissue Atlas (KPMP) Datasets. The source code and the Docker have been made publicly available at https://github.com/ddrrnn123/Omni-Seg.
CircleSnake: Instance Segmentation with Circle Representation
Nov 02, 2022Abstract:Circle representation has recently been introduced as a medical imaging optimized representation for more effective instance object detection on ball-shaped medical objects. With its superior performance on instance detection, it is appealing to extend the circle representation to instance medical object segmentation. In this work, we propose CircleSnake, a simple end-to-end circle contour deformation-based segmentation method for ball-shaped medical objects. Compared to the prevalent DeepSnake method, our contribution is three-fold: (1) We replace the complicated bounding box to octagon contour transformation with a computation-free and consistent bounding circle to circle contour adaption for segmenting ball-shaped medical objects; (2) Circle representation has fewer degrees of freedom (DoF=2) as compared with the octagon representation (DoF=8), thus yielding a more robust segmentation performance and better rotation consistency; (3) To the best of our knowledge, the proposed CircleSnake method is the first end-to-end circle representation deep segmentation pipeline method with consistent circle detection, circle contour proposal, and circular convolution. The key innovation is to integrate the circular graph convolution with circle detection into an end-to-end instance segmentation framework, enabled by the proposed simple and consistent circle contour representation. Glomeruli are used to evaluate the performance of the benchmarks. From the results, CircleSnake increases the average precision of glomerular detection from 0.559 to 0.614. The Dice score increased from 0.804 to 0.849. The code has been released: https://github.com/hrlblab/CircleSnake
Compound Figure Separation of Biomedical Images: Mining Large Datasets for Self-supervised Learning
Aug 30, 2022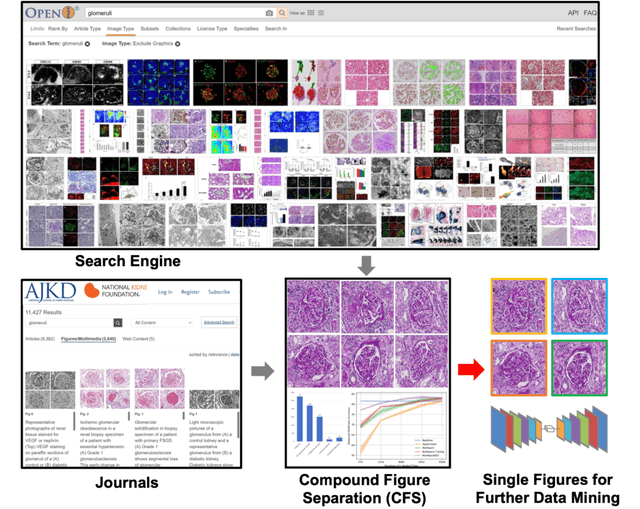
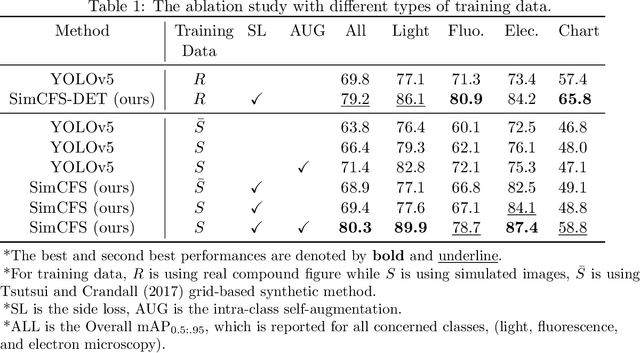
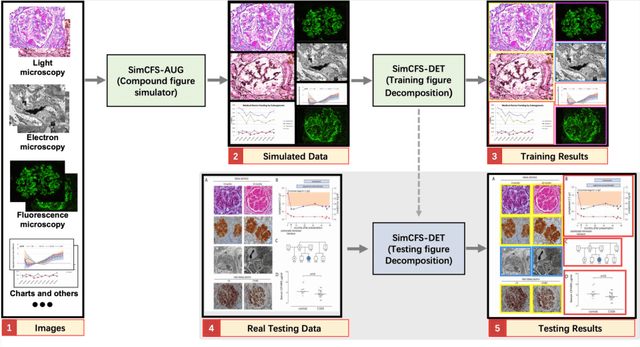
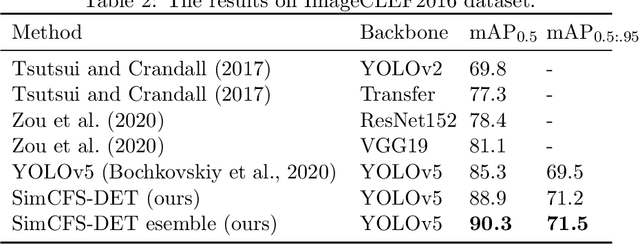
Abstract:With the rapid development of self-supervised learning (e.g., contrastive learning), the importance of having large-scale images (even without annotations) for training a more generalizable AI model has been widely recognized in medical image analysis. However, collecting large-scale task-specific unannotated data at scale can be challenging for individual labs. Existing online resources, such as digital books, publications, and search engines, provide a new resource for obtaining large-scale images. However, published images in healthcare (e.g., radiology and pathology) consist of a considerable amount of compound figures with subplots. In order to extract and separate compound figures into usable individual images for downstream learning, we propose a simple compound figure separation (SimCFS) framework without using the traditionally required detection bounding box annotations, with a new loss function and a hard case simulation. Our technical contribution is four-fold: (1) we introduce a simulation-based training framework that minimizes the need for resource extensive bounding box annotations; (2) we propose a new side loss that is optimized for compound figure separation; (3) we propose an intra-class image augmentation method to simulate hard cases; and (4) to the best of our knowledge, this is the first study that evaluates the efficacy of leveraging self-supervised learning with compound image separation. From the results, the proposed SimCFS achieved state-of-the-art performance on the ImageCLEF 2016 Compound Figure Separation Database. The pretrained self-supervised learning model using large-scale mined figures improved the accuracy of downstream image classification tasks with a contrastive learning algorithm. The source code of SimCFS is made publicly available at https://github.com/hrlblab/ImageSeperation.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://www.melba-journal.org/papers/2022:025.html. arXiv admin note: substantial text overlap with arXiv:2107.08650
Omni-Seg+: A Scale-aware Dynamic Network for Pathological Image Segmentation
Jun 27, 2022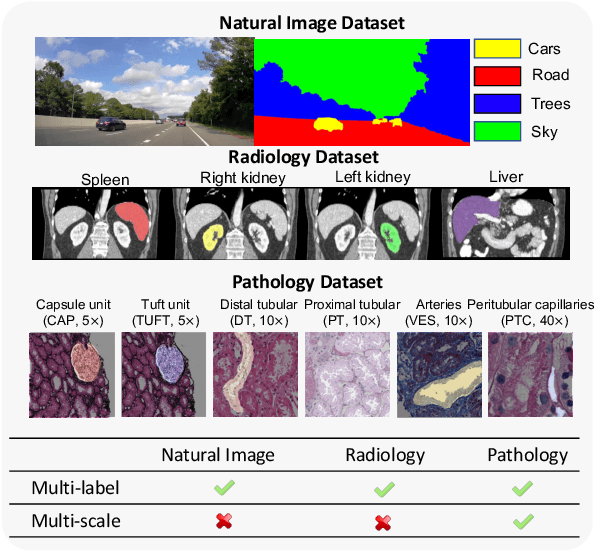
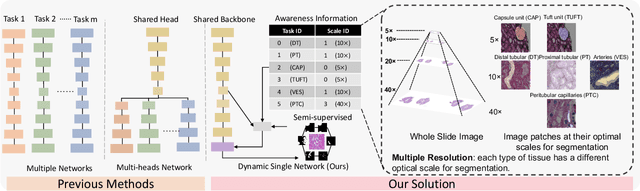
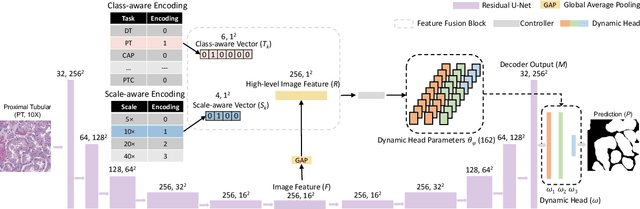
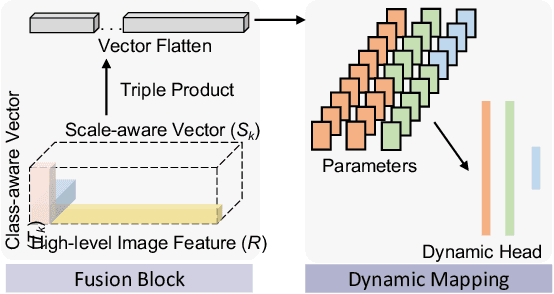
Abstract:Comprehensive semantic segmentation on renal pathological images is challenging due to the heterogeneous scales of the objects. For example, on a whole slide image (WSI), the cross-sectional areas of glomeruli can be 64 times larger than that of the peritubular capillaries, making it impractical to segment both objects on the same patch, at the same scale. To handle this scaling issue, prior studies have typically trained multiple segmentation networks in order to match the optimal pixel resolution of heterogeneous tissue types. This multi-network solution is resource-intensive and fails to model the spatial relationship between tissue types. In this paper, we propose the Omni-Seg+ network, a scale-aware dynamic neural network that achieves multi-object (six tissue types) and multi-scale (5X to 40X scale) pathological image segmentation via a single neural network. The contribution of this paper is three-fold: (1) a novel scale-aware controller is proposed to generalize the dynamic neural network from single-scale to multi-scale; (2) semi-supervised consistency regularization of pseudo-labels is introduced to model the inter-scale correlation of unannotated tissue types into a single end-to-end learning paradigm; and (3) superior scale-aware generalization is evidenced by directly applying a model trained on human kidney images to mouse kidney images, without retraining. By learning from ~150,000 human pathological image patches from six tissue types at three different resolutions, our approach achieved superior segmentation performance according to human visual assessment and evaluation of image-omics (i.e., spatial transcriptomics). The official implementation is available at https://github.com/ddrrnn123/Omni-Seg.
Glo-In-One: Holistic Glomerular Detection, Segmentation, and Lesion Characterization with Large-scale Web Image Mining
May 31, 2022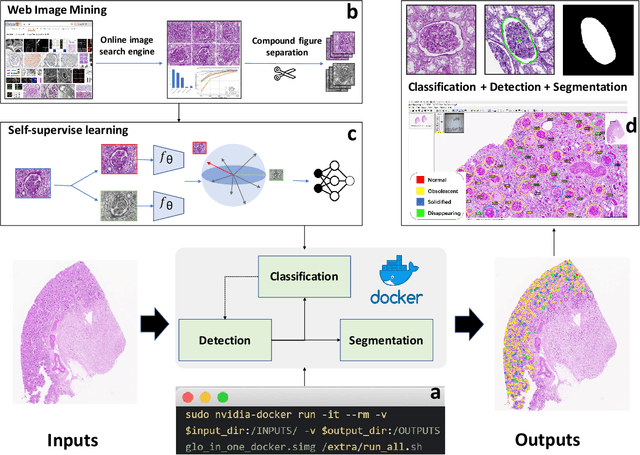
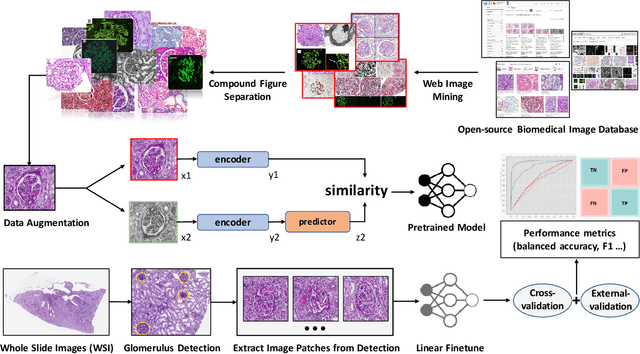
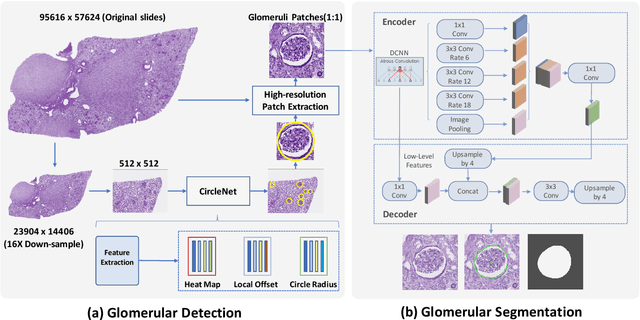
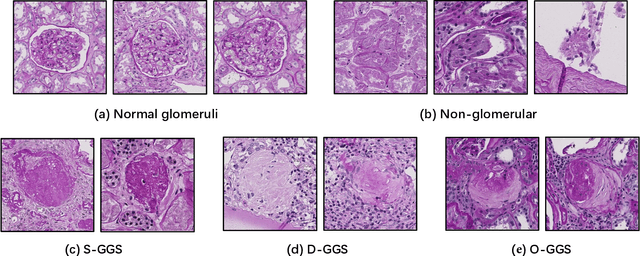
Abstract:The quantitative detection, segmentation, and characterization of glomeruli from high-resolution whole slide imaging (WSI) play essential roles in the computer-assisted diagnosis and scientific research in digital renal pathology. Historically, such comprehensive quantification requires extensive programming skills in order to be able to handle heterogeneous and customized computational tools. To bridge the gap of performing glomerular quantification for non-technical users, we develop the Glo-In-One toolkit to achieve holistic glomerular detection, segmentation, and characterization via a single line of command. Additionally, we release a large-scale collection of 30,000 unlabeled glomerular images to further facilitate the algorithmic development of self-supervised deep learning. The inputs of the Glo-In-One toolkit are WSIs, while the outputs are (1) WSI-level multi-class circle glomerular detection results (which can be directly manipulated with ImageScope), (2) glomerular image patches with segmentation masks, and (3) different lesion types. To leverage the performance of the Glo-In-One toolkit, we introduce self-supervised deep learning to glomerular quantification via large-scale web image mining. The GGS fine-grained classification model achieved a decent performance compared with baseline supervised methods while only using 10% of the annotated data. The glomerular detection achieved an average precision of 0.627 with circle representations, while the glomerular segmentation achieved a 0.955 patch-wise Dice Similarity Coefficient (DSC).
Deep Multi-modal Fusion of Image and Non-image Data in Disease Diagnosis and Prognosis: A Review
Mar 30, 2022



Abstract:The rapid development of diagnostic technologies in healthcare is leading to higher requirements for physicians to handle and integrate the heterogeneous, yet complementary data that are produced during routine practice. For instance, the personalized diagnosis and treatment planning for a single cancer patient relies on the various images (e.g., radiological, pathological, and camera images) and non-image data (e.g., clinical data and genomic data). However, such decision-making procedures can be subjective, qualitative, and have large inter-subject variabilities. With the recent advances in multi-modal deep learning technologies, an increasingly large number of efforts have been devoted to a key question: how do we extract and aggregate multi-modal information to ultimately provide more objective, quantitative computer-aided clinical decision making? This paper reviews the recent studies on dealing with such a question. Briefly, this review will include the (1) overview of current multi-modal learning workflows, (2) summarization of multi-modal fusion methods, (3) discussion of the performance, (4) applications in disease diagnosis and prognosis, and (5) challenges and future directions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge