Peize Li
Multimodal Hypothetical Summary for Retrieval-based Multi-image Question Answering
Dec 19, 2024Abstract:Retrieval-based multi-image question answering (QA) task involves retrieving multiple question-related images and synthesizing these images to generate an answer. Conventional "retrieve-then-answer" pipelines often suffer from cascading errors because the training objective of QA fails to optimize the retrieval stage. To address this issue, we propose a novel method to effectively introduce and reference retrieved information into the QA. Given the image set to be retrieved, we employ a multimodal large language model (visual perspective) and a large language model (textual perspective) to obtain multimodal hypothetical summary in question-form and description-form. By combining visual and textual perspectives, MHyS captures image content more specifically and replaces real images in retrieval, which eliminates the modality gap by transforming into text-to-text retrieval and helps improve retrieval. To more advantageously introduce retrieval with QA, we employ contrastive learning to align queries (questions) with MHyS. Moreover, we propose a coarse-to-fine strategy for calculating both sentence-level and word-level similarity scores, to further enhance retrieval and filter out irrelevant details. Our approach achieves a 3.7% absolute improvement over state-of-the-art methods on RETVQA and a 14.5% improvement over CLIP. Comprehensive experiments and detailed ablation studies demonstrate the superiority of our method.
Multimodal Indoor Localization Using Crowdsourced Radio Maps
Nov 17, 2023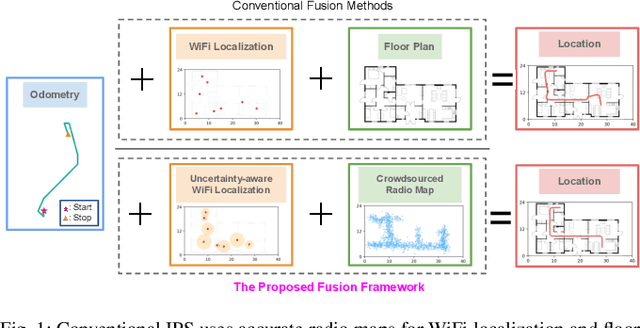
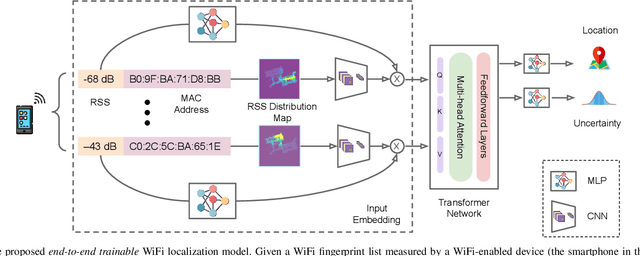
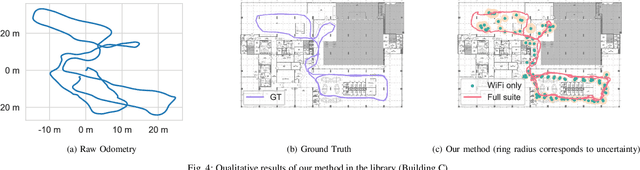
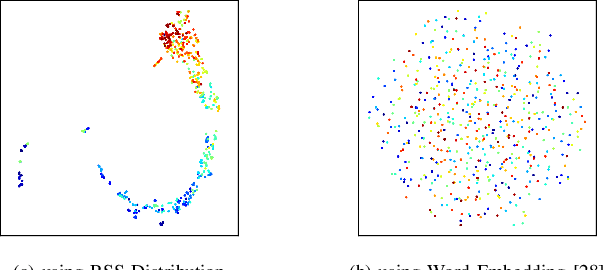
Abstract:Indoor Positioning Systems (IPS) traditionally rely on odometry and building infrastructures like WiFi, often supplemented by building floor plans for increased accuracy. However, the limitation of floor plans in terms of availability and timeliness of updates challenges their wide applicability. In contrast, the proliferation of smartphones and WiFi-enabled robots has made crowdsourced radio maps - databases pairing locations with their corresponding Received Signal Strengths (RSS) - increasingly accessible. These radio maps not only provide WiFi fingerprint-location pairs but encode movement regularities akin to the constraints imposed by floor plans. This work investigates the possibility of leveraging these radio maps as a substitute for floor plans in multimodal IPS. We introduce a new framework to address the challenges of radio map inaccuracies and sparse coverage. Our proposed system integrates an uncertainty-aware neural network model for WiFi localization and a bespoken Bayesian fusion technique for optimal fusion. Extensive evaluations on multiple real-world sites indicate a significant performance enhancement, with results showing ~ 25% improvement over the best baseline
Robust Human Detection under Visual Degradation via Thermal and mmWave Radar Fusion
Jul 07, 2023Abstract:The majority of human detection methods rely on the sensor using visible lights (e.g., RGB cameras) but such sensors are limited in scenarios with degraded vision conditions. In this paper, we present a multimodal human detection system that combines portable thermal cameras and single-chip mmWave radars. To mitigate the noisy detection features caused by the low contrast of thermal cameras and the multi-path noise of radar point clouds, we propose a Bayesian feature extractor and a novel uncertainty-guided fusion method that surpasses a variety of competing methods, either single-modal or multi-modal. We evaluate the proposed method on real-world data collection and demonstrate that our approach outperforms the state-of-the-art methods by a large margin.
Democratizing Pathological Image Segmentation with Lay Annotators via Molecular-empowered Learning
May 31, 2023Abstract:Multi-class cell segmentation in high-resolution Giga-pixel whole slide images (WSI) is critical for various clinical applications. Training such an AI model typically requires labor-intensive pixel-wise manual annotation from experienced domain experts (e.g., pathologists). Moreover, such annotation is error-prone when differentiating fine-grained cell types (e.g., podocyte and mesangial cells) via the naked human eye. In this study, we assess the feasibility of democratizing pathological AI deployment by only using lay annotators (annotators without medical domain knowledge). The contribution of this paper is threefold: (1) We proposed a molecular-empowered learning scheme for multi-class cell segmentation using partial labels from lay annotators; (2) The proposed method integrated Giga-pixel level molecular-morphology cross-modality registration, molecular-informed annotation, and molecular-oriented segmentation model, so as to achieve significantly superior performance via 3 lay annotators as compared with 2 experienced pathologists; (3) A deep corrective learning (learning with imperfect label) method is proposed to further improve the segmentation performance using partially annotated noisy data. From the experimental results, our learning method achieved F1 = 0.8496 using molecular-informed annotations from lay annotators, which is better than conventional morphology-based annotations (F1 = 0.7051) from experienced pathologists. Our method democratizes the development of a pathological segmentation deep model to the lay annotator level, which consequently scales up the learning process similar to a non-medical computer vision task. The official implementation and cell annotations are publicly available at https://github.com/hrlblab/MolecularEL.
An End-to-end Pipeline for 3D Slide-wise Multi-stain Renal Pathology Registration
May 19, 2023Abstract:Tissue examination and quantification in a 3D context on serial section whole slide images (WSIs) were laborintensive and time-consuming tasks. Our previous study proposed a novel registration-based method (Map3D) to automatically align WSIs to the same physical space, reducing the human efforts of screening serial sections from WSIs. However, the registration performance of our Map3D method was only evaluated on single-stain WSIs with large-scale kidney tissue samples. In this paper, we provide a Docker for an end-to-end 3D slide-wise registration pipeline on needle biopsy serial sections in a multi-stain paradigm. The contribution of this study is three-fold: (1) We release a containerized Docker for an end-to-end multi-stain WSI registration. (2) We prove that the Map3D pipeline is capable of sectional registration from multi-stain WSI. (3) We verify that the Map3D pipeline can also be applied to needle biopsy tissue samples. The source code and the Docker have been made publicly available at https://github.com/hrlblab/Map3D.
* 6 pages, 4 figures
OdomBeyondVision: An Indoor Multi-modal Multi-platform Odometry Dataset Beyond the Visible Spectrum
Jun 03, 2022
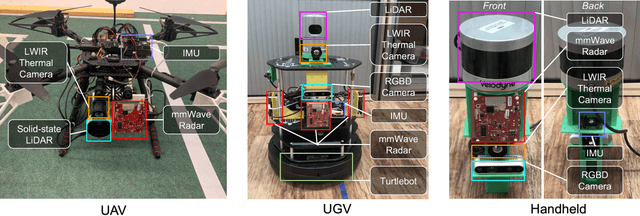


Abstract:This paper presents a multimodal indoor odometry dataset, OdomBeyondVision, featuring multiple sensors across the different spectrum and collected with different mobile platforms. Not only does OdomBeyondVision contain the traditional navigation sensors, sensors such as IMUs, mechanical LiDAR, RGBD camera, it also includes several emerging sensors such as the single-chip mmWave radar, LWIR thermal camera and solid-state LiDAR. With the above sensors on UAV, UGV and handheld platforms, we respectively recorded the multimodal odometry data and their movement trajectories in various indoor scenes and different illumination conditions. We release the exemplar radar, radar-inertial and thermal-inertial odometry implementations to demonstrate their results for future works to compare against and improve upon. The full dataset including toolkit and documentation is publicly available at: https://github.com/MAPS-Lab/OdomBeyondVision.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge