Aadarsh Jha
Compound Figure Separation of Biomedical Images: Mining Large Datasets for Self-supervised Learning
Aug 30, 2022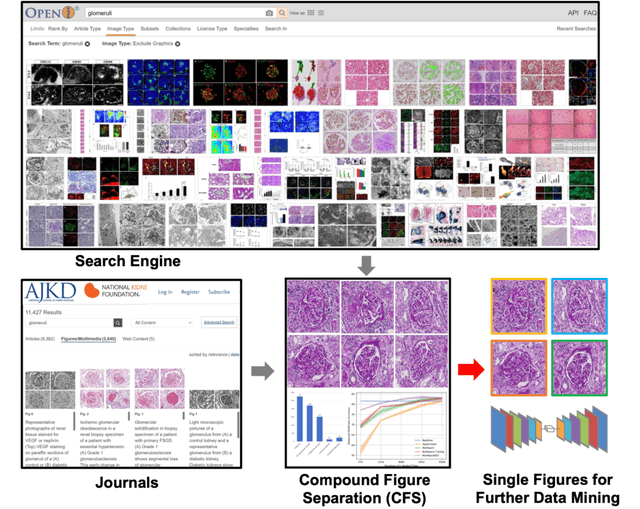
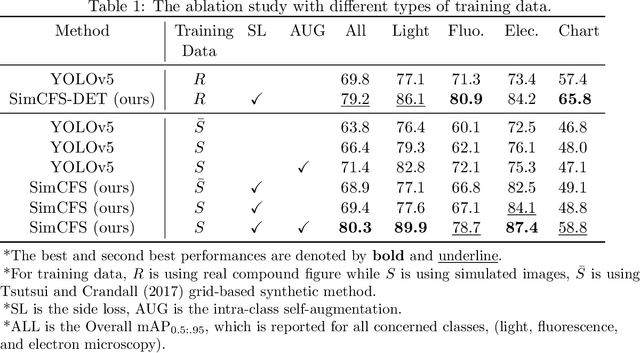
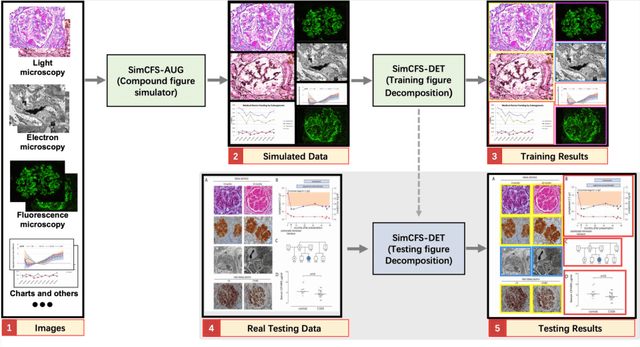
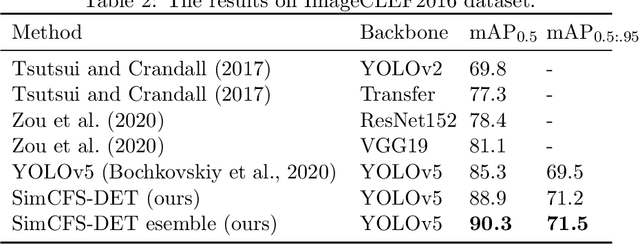
Abstract:With the rapid development of self-supervised learning (e.g., contrastive learning), the importance of having large-scale images (even without annotations) for training a more generalizable AI model has been widely recognized in medical image analysis. However, collecting large-scale task-specific unannotated data at scale can be challenging for individual labs. Existing online resources, such as digital books, publications, and search engines, provide a new resource for obtaining large-scale images. However, published images in healthcare (e.g., radiology and pathology) consist of a considerable amount of compound figures with subplots. In order to extract and separate compound figures into usable individual images for downstream learning, we propose a simple compound figure separation (SimCFS) framework without using the traditionally required detection bounding box annotations, with a new loss function and a hard case simulation. Our technical contribution is four-fold: (1) we introduce a simulation-based training framework that minimizes the need for resource extensive bounding box annotations; (2) we propose a new side loss that is optimized for compound figure separation; (3) we propose an intra-class image augmentation method to simulate hard cases; and (4) to the best of our knowledge, this is the first study that evaluates the efficacy of leveraging self-supervised learning with compound image separation. From the results, the proposed SimCFS achieved state-of-the-art performance on the ImageCLEF 2016 Compound Figure Separation Database. The pretrained self-supervised learning model using large-scale mined figures improved the accuracy of downstream image classification tasks with a contrastive learning algorithm. The source code of SimCFS is made publicly available at https://github.com/hrlblab/ImageSeperation.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://www.melba-journal.org/papers/2022:025.html. arXiv admin note: substantial text overlap with arXiv:2107.08650
Glo-In-One: Holistic Glomerular Detection, Segmentation, and Lesion Characterization with Large-scale Web Image Mining
May 31, 2022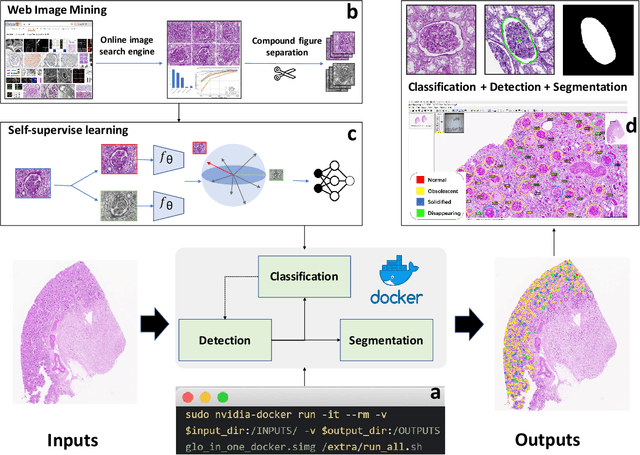
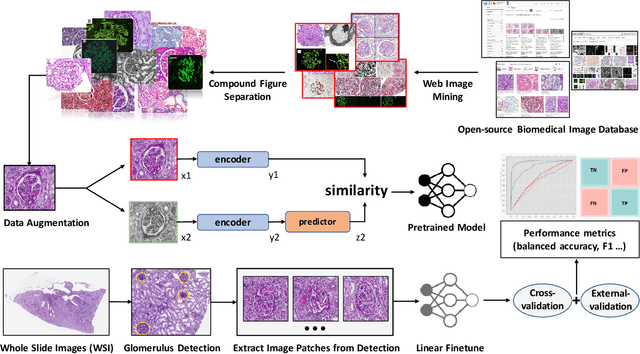
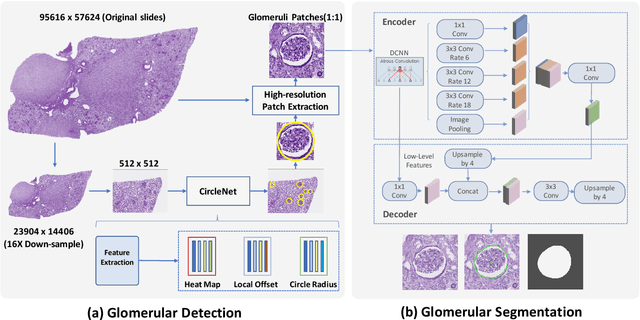
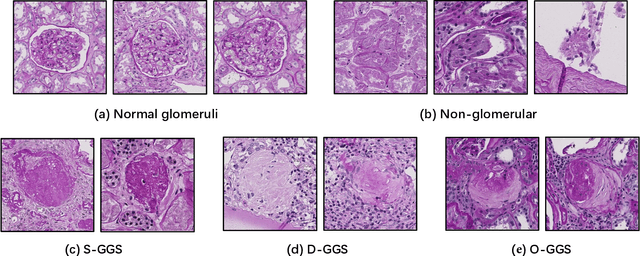
Abstract:The quantitative detection, segmentation, and characterization of glomeruli from high-resolution whole slide imaging (WSI) play essential roles in the computer-assisted diagnosis and scientific research in digital renal pathology. Historically, such comprehensive quantification requires extensive programming skills in order to be able to handle heterogeneous and customized computational tools. To bridge the gap of performing glomerular quantification for non-technical users, we develop the Glo-In-One toolkit to achieve holistic glomerular detection, segmentation, and characterization via a single line of command. Additionally, we release a large-scale collection of 30,000 unlabeled glomerular images to further facilitate the algorithmic development of self-supervised deep learning. The inputs of the Glo-In-One toolkit are WSIs, while the outputs are (1) WSI-level multi-class circle glomerular detection results (which can be directly manipulated with ImageScope), (2) glomerular image patches with segmentation masks, and (3) different lesion types. To leverage the performance of the Glo-In-One toolkit, we introduce self-supervised deep learning to glomerular quantification via large-scale web image mining. The GGS fine-grained classification model achieved a decent performance compared with baseline supervised methods while only using 10% of the annotated data. The glomerular detection achieved an average precision of 0.627 with circle representations, while the glomerular segmentation achieved a 0.955 patch-wise Dice Similarity Coefficient (DSC).
Compound Figure Separation of Biomedical Images with Side Loss
Jul 19, 2021



Abstract:Unsupervised learning algorithms (e.g., self-supervised learning, auto-encoder, contrastive learning) allow deep learning models to learn effective image representations from large-scale unlabeled data. In medical image analysis, even unannotated data can be difficult to obtain for individual labs. Fortunately, national-level efforts have been made to provide efficient access to obtain biomedical image data from previous scientific publications. For instance, NIH has launched the Open-i search engine that provides a large-scale image database with free access. However, the images in scientific publications consist of a considerable amount of compound figures with subplots. To extract and curate individual subplots, many different compound figure separation approaches have been developed, especially with the recent advances in deep learning. However, previous approaches typically required resource extensive bounding box annotation to train detection models. In this paper, we propose a simple compound figure separation (SimCFS) framework that uses weak classification annotations from individual images. Our technical contribution is three-fold: (1) we introduce a new side loss that is designed for compound figure separation; (2) we introduce an intra-class image augmentation method to simulate hard cases; (3) the proposed framework enables an efficient deployment to new classes of images, without requiring resource extensive bounding box annotations. From the results, the SimCFS achieved a new state-of-the-art performance on the ImageCLEF 2016 Compound Figure Separation Database. The source code of SimCFS is made publicly available at https://github.com/hrlblab/ImageSeperation.
VoxelEmbed: 3D Instance Segmentation and Tracking with Voxel Embedding based Deep Learning
Jun 22, 2021



Abstract:Recent advances in bioimaging have provided scientists a superior high spatial-temporal resolution to observe dynamics of living cells as 3D volumetric videos. Unfortunately, the 3D biomedical video analysis is lagging, impeded by resource insensitive human curation using off-the-shelf 3D analytic tools. Herein, biologists often need to discard a considerable amount of rich 3D spatial information by compromising on 2D analysis via maximum intensity projection. Recently, pixel embedding-based cell instance segmentation and tracking provided a neat and generalizable computing paradigm for understanding cellular dynamics. In this work, we propose a novel spatial-temporal voxel-embedding (VoxelEmbed) based learning method to perform simultaneous cell instance segmenting and tracking on 3D volumetric video sequences. Our contribution is in four-fold: (1) The proposed voxel embedding generalizes the pixel embedding with 3D context information; (2) Present a simple multi-stream learning approach that allows effective spatial-temporal embedding; (3) Accomplished an end-to-end framework for one-stage 3D cell instance segmentation and tracking without heavy parameter tuning; (4) The proposed 3D quantification is memory efficient via a single GPU with 12 GB memory. We evaluate our VoxelEmbed method on four 3D datasets (with different cell types) from the ISBI Cell Tracking Challenge. The proposed VoxelEmbed method achieved consistent superior overall performance (OP) on two densely annotated datasets. The performance is also competitive on two sparsely annotated cohorts with 20.6% and 2% of data-set having segmentation annotations. The results demonstrate that the VoxelEmbed method is a generalizable and memory-efficient solution.
SimTriplet: Simple Triplet Representation Learning with a Single GPU
Mar 09, 2021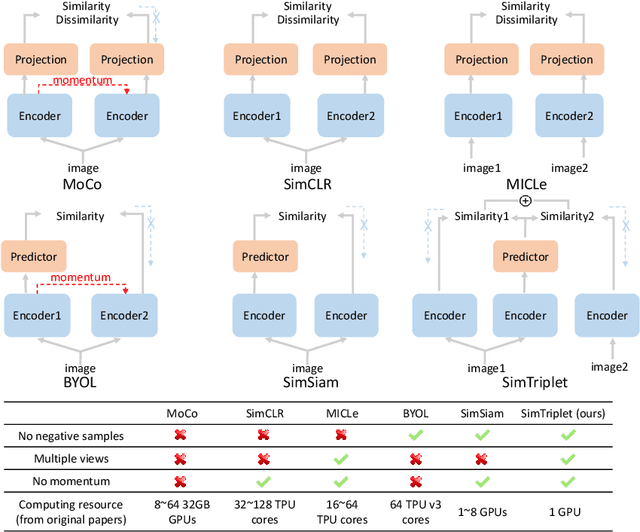
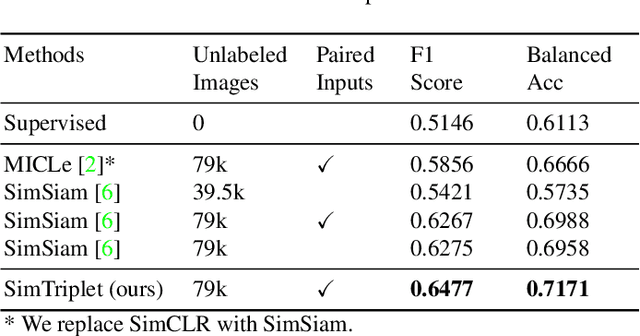
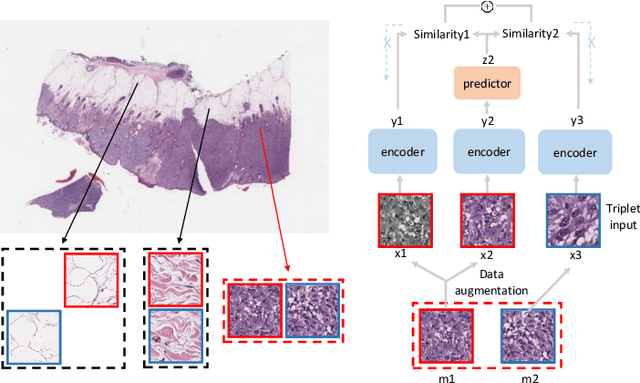
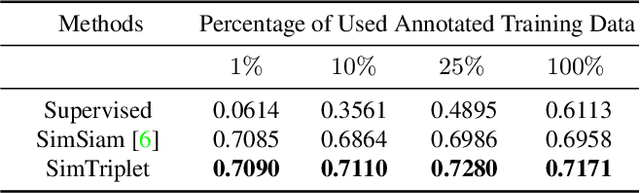
Abstract:Contrastive learning is a key technique of modern self-supervised learning. The broader accessibility of earlier approaches is hindered by the need of heavy computational resources (e.g., at least 8 GPUs or 32 TPU cores), which accommodate for large-scale negative samples or momentum. The more recent SimSiam approach addresses such key limitations via stop-gradient without momentum encoders. In medical image analysis, multiple instances can be achieved from the same patient or tissue. Inspired by these advances, we propose a simple triplet representation learning (SimTriplet) approach on pathological images. The contribution of the paper is three-fold: (1) The proposed SimTriplet method takes advantage of the multi-view nature of medical images beyond self-augmentation; (2) The method maximizes both intra-sample and inter-sample similarities via triplets from positive pairs, without using negative samples; and (3) The recent mix precision training is employed to advance the training by only using a single GPU with 16GB memory. By learning from 79,000 unlabeled pathological patch images, SimTriplet achieved 10.58% better performance compared with supervised learning. It also achieved 2.13% better performance compared with SimSiam. Our proposed SimTriplet can achieve decent performance using only 1% labeled data. The code and data are available at https://github.com/hrlblab/SimTriple.
Contrastive Learning Meets Transfer Learning: A Case Study In Medical Image Analysis
Mar 04, 2021



Abstract:Annotated medical images are typically rarer than labeled natural images since they are limited by domain knowledge and privacy constraints. Recent advances in transfer and contrastive learning have provided effective solutions to tackle such issues from different perspectives. The state-of-the-art transfer learning (e.g., Big Transfer (BiT)) and contrastive learning (e.g., Simple Siamese Contrastive Learning (SimSiam)) approaches have been investigated independently, without considering the complementary nature of such techniques. It would be appealing to accelerate contrastive learning with transfer learning, given that slow convergence speed is a critical limitation of modern contrastive learning approaches. In this paper, we investigate the feasibility of aligning BiT with SimSiam. From empirical analyses, different normalization techniques (Group Norm in BiT vs. Batch Norm in SimSiam) are the key hurdle of adapting BiT to SimSiam. When combining BiT with SimSiam, we evaluated the performance of using BiT, SimSiam, and BiT+SimSiam on CIFAR-10 and HAM10000 datasets. The results suggest that the BiT models accelerate the convergence speed of SimSiam. When used together, the model gives superior performance over both of its counterparts. We hope this study will motivate researchers to revisit the task of aggregating big pre-trained models with contrastive learning models for image analysis.
BEDS: Bagging ensemble deep segmentation for nucleus segmentation with testing stage stain augmentation
Feb 17, 2021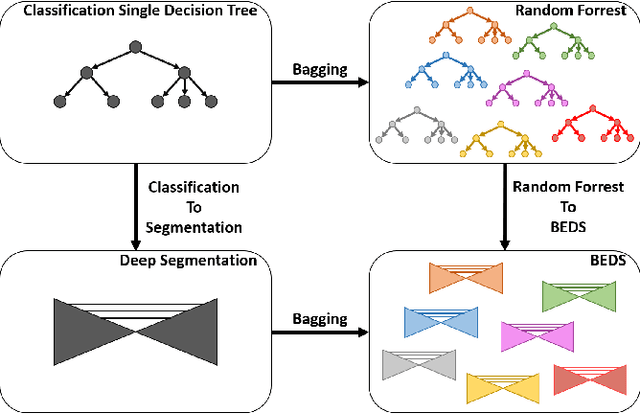
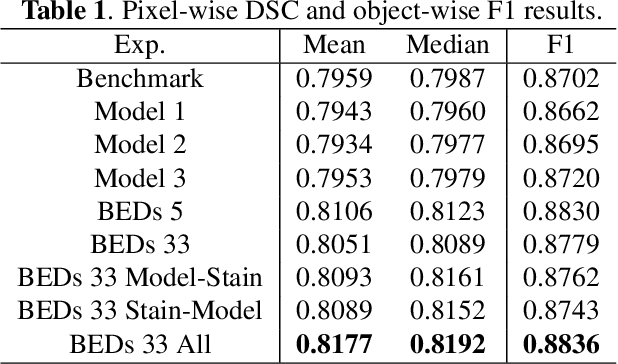
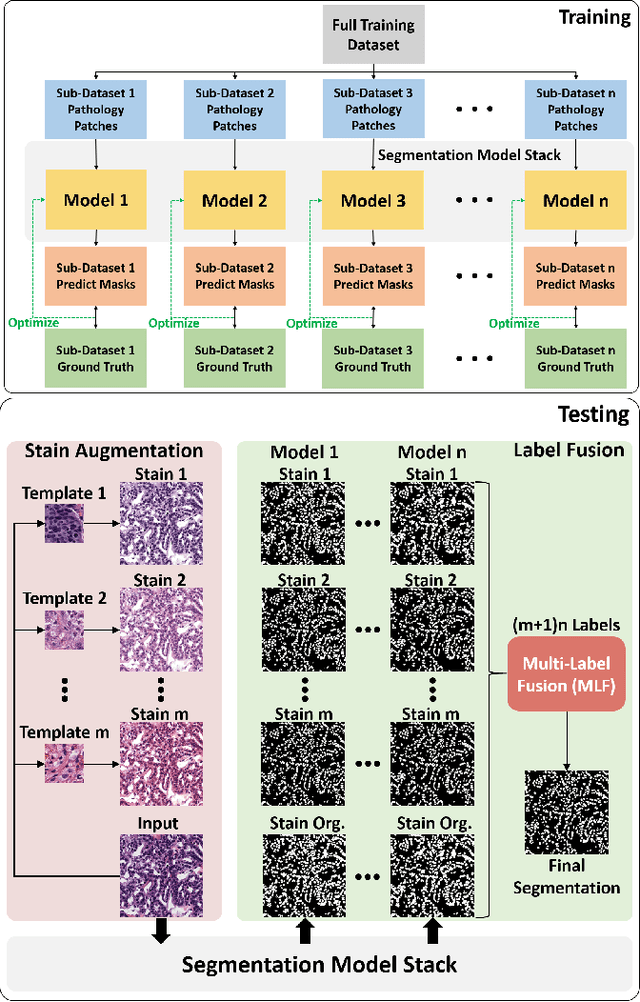

Abstract:Reducing outcome variance is an essential task in deep learning based medical image analysis. Bootstrap aggregating, also known as bagging, is a canonical ensemble algorithm for aggregating weak learners to become a strong learner. Random forest is one of the most powerful machine learning algorithms before deep learning era, whose superior performance is driven by fitting bagged decision trees (weak learners). Inspired by the random forest technique, we propose a simple bagging ensemble deep segmentation (BEDs) method to train multiple U-Nets with partial training data to segment dense nuclei on pathological images. The contributions of this study are three-fold: (1) developing a self-ensemble learning framework for nucleus segmentation; (2) aggregating testing stage augmentation with self-ensemble learning; and (3) elucidating the idea that self-ensemble and testing stage stain augmentation are complementary strategies for a superior segmentation performance. Implementation Detail: https://github.com/xingli1102/BEDs.
Towards Annotation-free Instance Segmentation and Tracking with Adversarial Simulations
Jan 19, 2021



Abstract:The quantitative analysis of microscope videos often requires instance segmentation and tracking of cellular and subcellular objects. The traditional method is composed of two stages: (1) performing instance object segmentation of each frame, and (2) associating objects frame-by-frame. Recently, pixel-embedding-based deep learning approaches provide single stage holistic solutions to tackle instance segmentation and tracking simultaneously. However, such deep learning methods require consistent annotations not only spatially (for segmentation), but also temporally (for tracking). In computer vision, annotated training data with consistent segmentation and tracking is resource intensive, the severity of which can be multiplied in microscopy imaging due to (1) dense objects (e.g., overlapping or touching), and (2) high dynamics (e.g., irregular motion and mitosis). To alleviate the lack of such annotations in dynamics scenes, adversarial simulations have provided successful solutions in computer vision, such as using simulated environments (e.g., computer games) to train real-world self-driving systems. In this paper, we propose an annotation-free synthetic instance segmentation and tracking (ASIST) method with adversarial simulation and single-stage pixel-embedding based learning. The contribution of this paper is three-fold: (1) the proposed method aggregates adversarial simulations and single-stage pixel-embedding based deep learning; (2) the method is assessed with both the cellular (i.e., HeLa cells) and subcellular (i.e., microvilli) objects; and (3) to the best of our knowledge, this is the first study to explore annotation-free instance segmentation and tracking study for microscope videos. This ASIST method achieved an important step forward, when compared with fully supervised approaches.
ASIST: Annotation-free synthetic instance segmentation and tracking for microscope video analysis
Nov 02, 2020



Abstract:Instance object segmentation and tracking provide comprehensive quantification of objects across microscope videos. The recent single-stage pixel-embedding based deep learning approach has shown its superior performance compared with "segment-then-associate" two-stage solutions. However, one major limitation of applying a supervised pixel-embedding based method to microscope videos is the resource-intensive manual labeling, which involves tracing hundreds of overlapped objects with their temporal associations across video frames. Inspired by the recent generative adversarial network (GAN) based annotation-free image segmentation, we propose a novel annotation-free synthetic instance segmentation and tracking (ASIST) algorithm for analyzing microscope videos of sub-cellular microvilli. The contributions of this paper are three-fold: (1) proposing a new annotation-free video analysis paradigm is proposed. (2) aggregating the embedding based instance segmentation and tracking with annotation-free synthetic learning as a holistic framework; and (3) to the best of our knowledge, this is first study to investigate microvilli instance segmentation and tracking using embedding based deep learning. From the experimental results, the proposed annotation-free method achieved superior performance compared with supervised learning.
CaCL: Class-aware Codebook Learning for Weakly Supervised Segmentation on Diffuse Image Patterns
Nov 02, 2020



Abstract:Weakly supervised learning has been rapidly advanced in biomedical image analysis to achieve pixel-wise labels (segmentation) from image-wise annotations (classification), as biomedical images naturally contain image-wise labels in many scenarios. The current weakly supervised learning algorithms from the computer vision community are largely designed for focal objects (e.g., dogs and cats). However, such algorithms are not optimized for diffuse patterns in biomedical imaging (e.g., stains and fluorescent in microscopy imaging). In this paper, we propose a novel class-aware codebook learning (CaCL) algorithm to perform weakly supervised learning for diffuse image patterns. Specifically, the CaCL algorithm is deployed to segment protein expressed brush border regions from histological images of human duodenum. This paper makes the following contributions: (1) we approach the weakly supervised segmentation from a novel codebook learning perspective; (2) the CaCL algorithm segments diffuse image patterns rather than focal objects; and (3) The proposed algorithm is implemented in a multi-task framework based on Vector Quantised-Variational AutoEncoder (VQ-VAE) to perform image reconstruction, classification, feature embedding, and segmentation. The experimental results show that our method achieved superior performance compared with baseline weakly supervised algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge