Catie Chang
Defining and Benchmarking a Data-Centric Design Space for Brain Graph Construction
Aug 17, 2025Abstract:The construction of brain graphs from functional Magnetic Resonance Imaging (fMRI) data plays a crucial role in enabling graph machine learning for neuroimaging. However, current practices often rely on rigid pipelines that overlook critical data-centric choices in how brain graphs are constructed. In this work, we adopt a Data-Centric AI perspective and systematically define and benchmark a data-centric design space for brain graph construction, constrasting with primarily model-centric prior work. We organize this design space into three stages: temporal signal processing, topology extraction, and graph featurization. Our contributions lie less in novel components and more in evaluating how combinations of existing and modified techniques influence downstream performance. Specifically, we study high-amplitude BOLD signal filtering, sparsification and unification strategies for connectivity, alternative correlation metrics, and multi-view node and edge features, such as incorporating lagged dynamics. Experiments on the HCP1200 and ABIDE datasets show that thoughtful data-centric configurations consistently improve classification accuracy over standard pipelines. These findings highlight the critical role of upstream data decisions and underscore the importance of systematically exploring the data-centric design space for graph-based neuroimaging. Our code is available at https://github.com/GeQinwen/DataCentricBrainGraphs.
Linear Spherical Sliced Optimal Transport: A Fast Metric for Comparing Spherical Data
Nov 09, 2024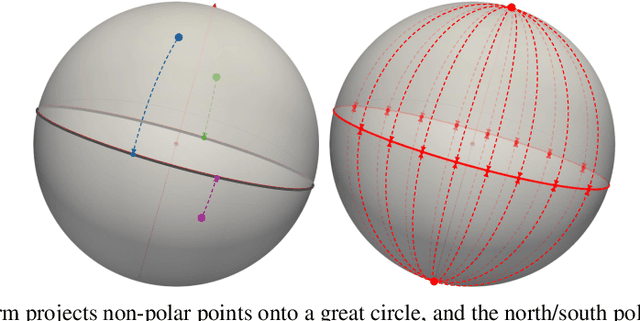
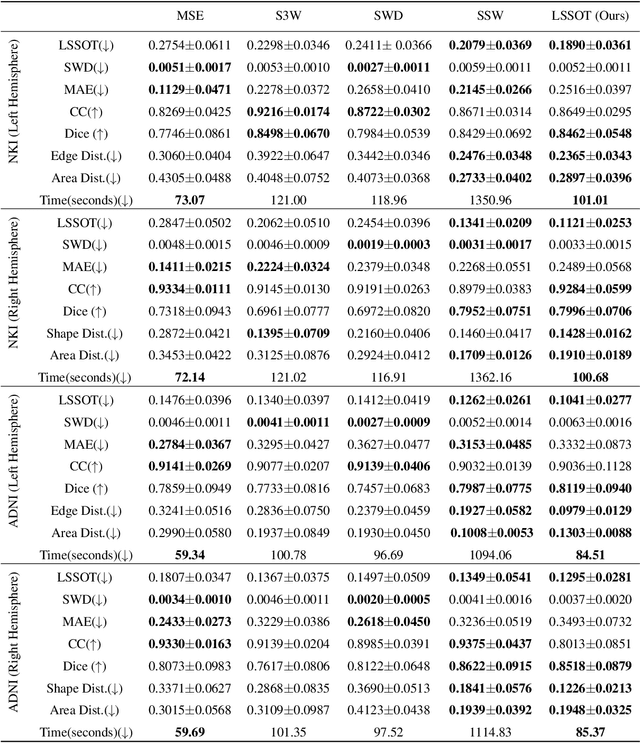
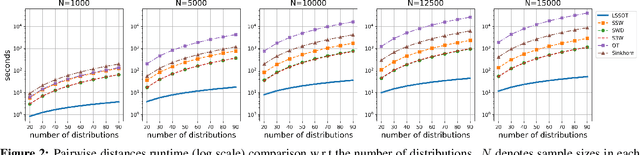
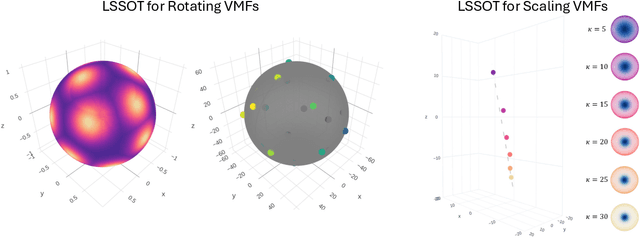
Abstract:Efficient comparison of spherical probability distributions becomes important in fields such as computer vision, geosciences, and medicine. Sliced optimal transport distances, such as spherical and stereographic spherical sliced Wasserstein distances, have recently been developed to address this need. These methods reduce the computational burden of optimal transport by slicing hyperspheres into one-dimensional projections, i.e., lines or circles. Concurrently, linear optimal transport has been proposed to embed distributions into \( L^2 \) spaces, where the \( L^2 \) distance approximates the optimal transport distance, thereby simplifying comparisons across multiple distributions. In this work, we introduce the Linear Spherical Sliced Optimal Transport (LSSOT) framework, which utilizes slicing to embed spherical distributions into \( L^2 \) spaces while preserving their intrinsic geometry, offering a computationally efficient metric for spherical probability measures. We establish the metricity of LSSOT and demonstrate its superior computational efficiency in applications such as cortical surface registration, 3D point cloud interpolation via gradient flow, and shape embedding. Our results demonstrate the significant computational benefits and high accuracy of LSSOT in these applications.
How Good Are We? Evaluating Cell AI Foundation Models in Kidney Pathology with Human-in-the-Loop Enrichment
Oct 31, 2024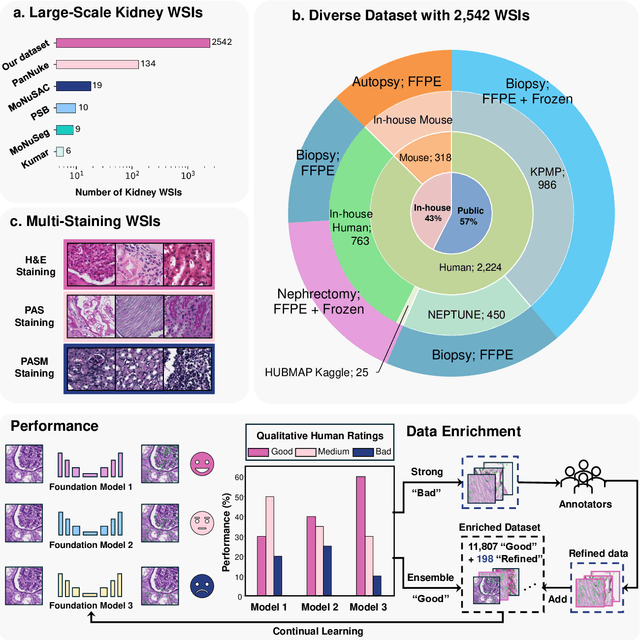
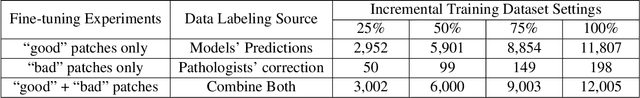
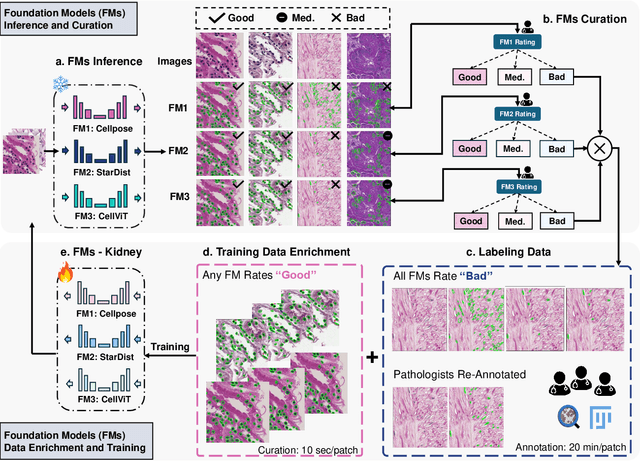
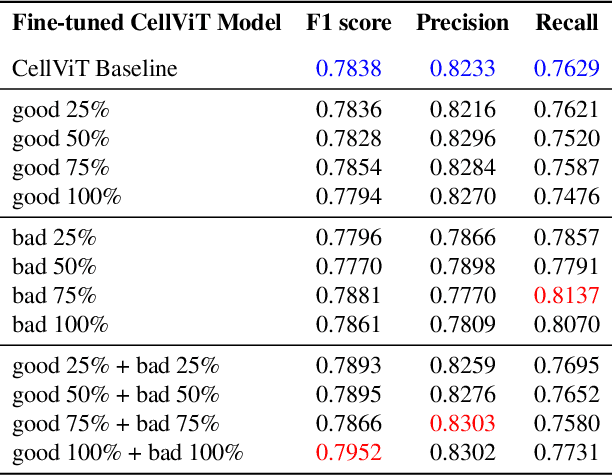
Abstract:Training AI foundation models has emerged as a promising large-scale learning approach for addressing real-world healthcare challenges, including digital pathology. While many of these models have been developed for tasks like disease diagnosis and tissue quantification using extensive and diverse training datasets, their readiness for deployment on some arguably simplest tasks, such as nuclei segmentation within a single organ (e.g., the kidney), remains uncertain. This paper seeks to answer this key question, "How good are we?", by thoroughly evaluating the performance of recent cell foundation models on a curated multi-center, multi-disease, and multi-species external testing dataset. Additionally, we tackle a more challenging question, "How can we improve?", by developing and assessing human-in-the-loop data enrichment strategies aimed at enhancing model performance while minimizing the reliance on pixel-level human annotation. To address the first question, we curated a multicenter, multidisease, and multispecies dataset consisting of 2,542 kidney whole slide images (WSIs). Three state-of-the-art (SOTA) cell foundation models-Cellpose, StarDist, and CellViT-were selected for evaluation. To tackle the second question, we explored data enrichment algorithms by distilling predictions from the different foundation models with a human-in-the-loop framework, aiming to further enhance foundation model performance with minimal human efforts. Our experimental results showed that all three foundation models improved over their baselines with model fine-tuning with enriched data. Interestingly, the baseline model with the highest F1 score does not yield the best segmentation outcomes after fine-tuning. This study establishes a benchmark for the development and deployment of cell vision foundation models tailored for real-world data applications.
NeuroBOLT: Resting-state EEG-to-fMRI Synthesis with Multi-dimensional Feature Mapping
Oct 07, 2024



Abstract:Functional magnetic resonance imaging (fMRI) is an indispensable tool in modern neuroscience, providing a non-invasive window into whole-brain dynamics at millimeter-scale spatial resolution. However, fMRI is constrained by issues such as high operation costs and immobility. With the rapid advancements in cross-modality synthesis and brain decoding, the use of deep neural networks has emerged as a promising solution for inferring whole-brain, high-resolution fMRI features directly from electroencephalography (EEG), a more widely accessible and portable neuroimaging modality. Nonetheless, the complex projection from neural activity to fMRI hemodynamic responses and the spatial ambiguity of EEG pose substantial challenges both in modeling and interpretability. Relatively few studies to date have developed approaches for EEG-fMRI translation, and although they have made significant strides, the inference of fMRI signals in a given study has been limited to a small set of brain areas and to a single condition (i.e., either resting-state or a specific task). The capability to predict fMRI signals in other brain areas, as well as to generalize across conditions, remain critical gaps in the field. To tackle these challenges, we introduce a novel and generalizable framework: NeuroBOLT, i.e., Neuro-to-BOLD Transformer, which leverages multi-dimensional representation learning from temporal, spatial, and spectral domains to translate raw EEG data to the corresponding fMRI activity signals across the brain. Our experiments demonstrate that NeuroBOLT effectively reconstructs resting-state fMRI signals from primary sensory, high-level cognitive areas, and deep subcortical brain regions, achieving state-of-the-art accuracy and significantly advancing the integration of these two modalities.
Reconstructing physiological signals from fMRI across the adult lifespan
Aug 26, 2024



Abstract:Interactions between the brain and body are of fundamental importance for human behavior and health. Functional magnetic resonance imaging (fMRI) captures whole-brain activity noninvasively, and modeling how fMRI signals interact with physiological dynamics of the body can provide new insight into brain function and offer potential biomarkers of disease. However, physiological recordings are not always possible to acquire since they require extra equipment and setup, and even when they are, the recorded physiological signals may contain substantial artifacts. To overcome this limitation, machine learning models have been proposed to directly extract features of respiratory and cardiac activity from resting-state fMRI signals. To date, such work has been carried out only in healthy young adults and in a pediatric population, leaving open questions about the efficacy of these approaches on older adults. Here, we propose a novel framework that leverages Transformer-based architectures for reconstructing two key physiological signals - low-frequency respiratory volume (RV) and heart rate (HR) fluctuations - from fMRI data, and test these models on a dataset of individuals aged 36-89 years old. Our framework outperforms previously proposed approaches (attaining median correlations between predicted and measured signals of r ~ .698 for RV and r ~ .618 for HR), indicating the potential of leveraging attention mechanisms to model fMRI-physiological signal relationships. We also evaluate several model training and fine-tuning strategies, and find that incorporating young-adult data during training improves the performance when predicting physiological signals in the aging cohort. Overall, our approach successfully infers key physiological variables directly from fMRI data from individuals across a wide range of the adult lifespan.
Assessment of Cell Nuclei AI Foundation Models in Kidney Pathology
Aug 09, 2024Abstract:Cell nuclei instance segmentation is a crucial task in digital kidney pathology. Traditional automatic segmentation methods often lack generalizability when applied to unseen datasets. Recently, the success of foundation models (FMs) has provided a more generalizable solution, potentially enabling the segmentation of any cell type. In this study, we perform a large-scale evaluation of three widely used state-of-the-art (SOTA) cell nuclei foundation models (Cellpose, StarDist, and CellViT). Specifically, we created a highly diverse evaluation dataset consisting of 2,542 kidney whole slide images (WSIs) collected from both human and rodent sources, encompassing various tissue types, sizes, and staining methods. To our knowledge, this is the largest-scale evaluation of its kind to date. Our quantitative analysis of the prediction distribution reveals a persistent performance gap in kidney pathology. Among the evaluated models, CellViT demonstrated superior performance in segmenting nuclei in kidney pathology. However, none of the foundation models are perfect; a performance gap remains in general nuclei segmentation for kidney pathology.
Leveraging sinusoidal representation networks to predict fMRI signals from EEG
Nov 06, 2023Abstract:In modern neuroscience, functional magnetic resonance imaging (fMRI) has been a crucial and irreplaceable tool that provides a non-invasive window into the dynamics of whole-brain activity. Nevertheless, fMRI is limited by hemodynamic blurring as well as high cost, immobility, and incompatibility with metal implants. Electroencephalography (EEG) is complementary to fMRI and can directly record the cortical electrical activity at high temporal resolution, but has more limited spatial resolution and is unable to recover information about deep subcortical brain structures. The ability to obtain fMRI information from EEG would enable cost-effective, imaging across a wider set of brain regions. Further, beyond augmenting the capabilities of EEG, cross-modality models would facilitate the interpretation of fMRI signals. However, as both EEG and fMRI are high-dimensional and prone to artifacts, it is currently challenging to model fMRI from EEG. To address this challenge, we propose a novel architecture that can predict fMRI signals directly from multi-channel EEG without explicit feature engineering. Our model achieves this by implementing a Sinusoidal Representation Network (SIREN) to learn frequency information in brain dynamics from EEG, which serves as the input to a subsequent encoder-decoder to effectively reconstruct the fMRI signal from a specific brain region. We evaluate our model using a simultaneous EEG-fMRI dataset with 8 subjects and investigate its potential for predicting subcortical fMRI signals. The present results reveal that our model outperforms a recent state-of-the-art model, and indicates the potential of leveraging periodic activation functions in deep neural networks to model functional neuroimaging data.
NeuroGraph: Benchmarks for Graph Machine Learning in Brain Connectomics
Jun 25, 2023Abstract:Machine learning provides a valuable tool for analyzing high-dimensional functional neuroimaging data, and is proving effective in predicting various neurological conditions, psychiatric disorders, and cognitive patterns. In functional Magnetic Resonance Imaging (MRI) research, interactions between brain regions are commonly modeled using graph-based representations. The potency of graph machine learning methods has been established across myriad domains, marking a transformative step in data interpretation and predictive modeling. Yet, despite their promise, the transposition of these techniques to the neuroimaging domain remains surprisingly under-explored due to the expansive preprocessing pipeline and large parameter search space for graph-based datasets construction. In this paper, we introduce NeuroGraph, a collection of graph-based neuroimaging datasets that span multiple categories of behavioral and cognitive traits. We delve deeply into the dataset generation search space by crafting 35 datasets within both static and dynamic contexts, running in excess of 15 baseline methods for benchmarking. Additionally, we provide generic frameworks for learning on dynamic as well as static graphs. Our extensive experiments lead to several key observations. Notably, using correlation vectors as node features, incorporating larger number of regions of interest, and employing sparser graphs lead to improved performance. To foster further advancements in graph-based data driven Neuroimaging, we offer a comprehensive open source Python package that includes the datasets, baseline implementations, model training, and standard evaluation. The package is publicly accessible at https://anwar-said.github.io/anwarsaid/neurograph.html .
Compound Figure Separation of Biomedical Images: Mining Large Datasets for Self-supervised Learning
Aug 30, 2022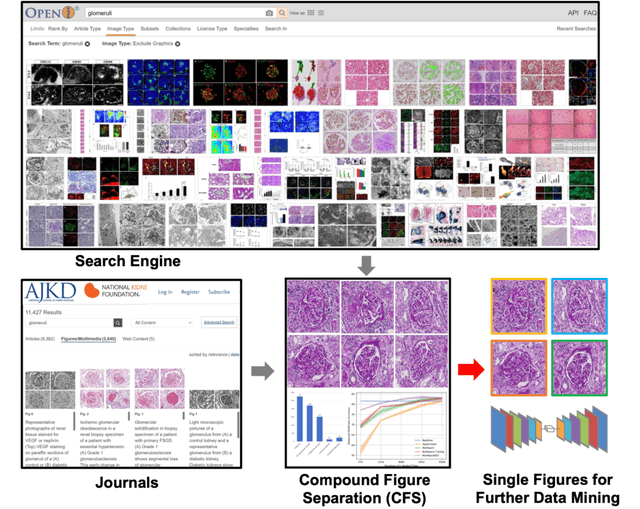
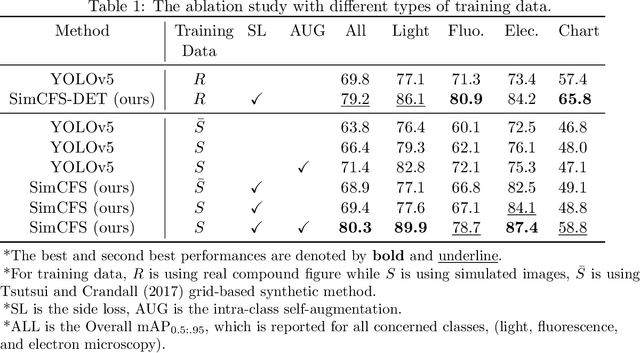
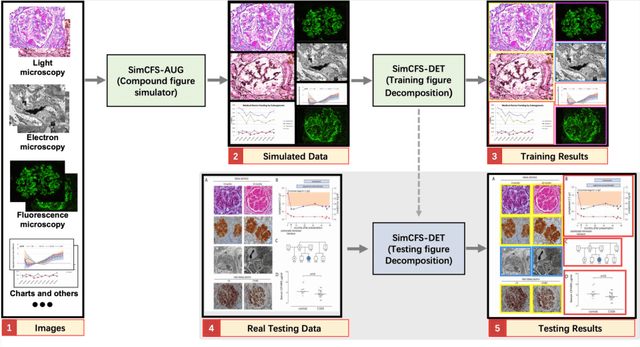
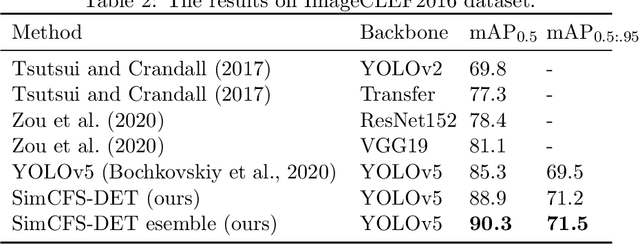
Abstract:With the rapid development of self-supervised learning (e.g., contrastive learning), the importance of having large-scale images (even without annotations) for training a more generalizable AI model has been widely recognized in medical image analysis. However, collecting large-scale task-specific unannotated data at scale can be challenging for individual labs. Existing online resources, such as digital books, publications, and search engines, provide a new resource for obtaining large-scale images. However, published images in healthcare (e.g., radiology and pathology) consist of a considerable amount of compound figures with subplots. In order to extract and separate compound figures into usable individual images for downstream learning, we propose a simple compound figure separation (SimCFS) framework without using the traditionally required detection bounding box annotations, with a new loss function and a hard case simulation. Our technical contribution is four-fold: (1) we introduce a simulation-based training framework that minimizes the need for resource extensive bounding box annotations; (2) we propose a new side loss that is optimized for compound figure separation; (3) we propose an intra-class image augmentation method to simulate hard cases; and (4) to the best of our knowledge, this is the first study that evaluates the efficacy of leveraging self-supervised learning with compound image separation. From the results, the proposed SimCFS achieved state-of-the-art performance on the ImageCLEF 2016 Compound Figure Separation Database. The pretrained self-supervised learning model using large-scale mined figures improved the accuracy of downstream image classification tasks with a contrastive learning algorithm. The source code of SimCFS is made publicly available at https://github.com/hrlblab/ImageSeperation.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://www.melba-journal.org/papers/2022:025.html. arXiv admin note: substantial text overlap with arXiv:2107.08650
Capture Agent Free Biosensing using Porous Silicon Arrays and Machine Learning
Jan 22, 2022



Abstract:Biosensors are an essential tool for medical diagnostics, environmental monitoring and food safety. Typically, biosensors are designed to detect specific analytes through functionalization with the appropriate capture agents. However, the use of capture agents limits the number of analytes that can be simultaneously detected and reduces the robustness of the biosensor. In this work, we report a versatile, capture agent free biosensor platform based on an array of porous silicon (PSi) thin films, which has the potential to robustly detect a wide variety of analytes based on their physical and chemical properties in the nanoscale porous media. The ability of this system to reproducibly classify, quantify, and discriminate three proteins is demonstrated to concentrations down to at least 0.02g/L (between 300nM and 450nM) by utilizing PSi array elements with a unique combination of pore size and buffer pH, employing linear discriminant analysis for dimensionality reduction, and using support vector machines as a classifier. This approach represents a significant step towards a low cost, simple and robust biosensor platform that is able to detect a vast range of biomolecules.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge