Yamin Li
DaRePlane: Direction-aware Representations for Dynamic Scene Reconstruction
Oct 18, 2024Abstract:Numerous recent approaches to modeling and re-rendering dynamic scenes leverage plane-based explicit representations, addressing slow training times associated with models like neural radiance fields (NeRF) and Gaussian splatting (GS). However, merely decomposing 4D dynamic scenes into multiple 2D plane-based representations is insufficient for high-fidelity re-rendering of scenes with complex motions. In response, we present DaRePlane, a novel direction-aware representation approach that captures scene dynamics from six different directions. This learned representation undergoes an inverse dual-tree complex wavelet transformation (DTCWT) to recover plane-based information. Within NeRF pipelines, DaRePlane computes features for each space-time point by fusing vectors from these recovered planes, then passed to a tiny MLP for color regression. When applied to Gaussian splatting, DaRePlane computes the features of Gaussian points, followed by a tiny multi-head MLP for spatial-time deformation prediction. Notably, to address redundancy introduced by the six real and six imaginary direction-aware wavelet coefficients, we introduce a trainable masking approach, mitigating storage issues without significant performance decline. To demonstrate the generality and efficiency of DaRePlane, we test it on both regular and surgical dynamic scenes, for both NeRF and GS systems. Extensive experiments show that DaRePlane yields state-of-the-art performance in novel view synthesis for various complex dynamic scenes.
Surgical Depth Anything: Depth Estimation for Surgical Scenes using Foundation Models
Oct 09, 2024


Abstract:Monocular depth estimation is crucial for tracking and reconstruction algorithms, particularly in the context of surgical videos. However, the inherent challenges in directly obtaining ground truth depth maps during surgery render supervised learning approaches impractical. While many self-supervised methods based on Structure from Motion (SfM) have shown promising results, they rely heavily on high-quality camera motion and require optimization on a per-patient basis. These limitations can be mitigated by leveraging the current state-of-the-art foundational model for depth estimation, Depth Anything. However, when directly applied to surgical scenes, Depth Anything struggles with issues such as blurring, bleeding, and reflections, resulting in suboptimal performance. This paper presents a fine-tuning of the Depth Anything model specifically for the surgical domain, aiming to deliver more accurate pixel-wise depth maps tailored to the unique requirements and challenges of surgical environments. Our fine-tuning approach significantly improves the model's performance in surgical scenes, reducing errors related to blurring and reflections, and achieving a more reliable and precise depth estimation.
NeuroBOLT: Resting-state EEG-to-fMRI Synthesis with Multi-dimensional Feature Mapping
Oct 07, 2024



Abstract:Functional magnetic resonance imaging (fMRI) is an indispensable tool in modern neuroscience, providing a non-invasive window into whole-brain dynamics at millimeter-scale spatial resolution. However, fMRI is constrained by issues such as high operation costs and immobility. With the rapid advancements in cross-modality synthesis and brain decoding, the use of deep neural networks has emerged as a promising solution for inferring whole-brain, high-resolution fMRI features directly from electroencephalography (EEG), a more widely accessible and portable neuroimaging modality. Nonetheless, the complex projection from neural activity to fMRI hemodynamic responses and the spatial ambiguity of EEG pose substantial challenges both in modeling and interpretability. Relatively few studies to date have developed approaches for EEG-fMRI translation, and although they have made significant strides, the inference of fMRI signals in a given study has been limited to a small set of brain areas and to a single condition (i.e., either resting-state or a specific task). The capability to predict fMRI signals in other brain areas, as well as to generalize across conditions, remain critical gaps in the field. To tackle these challenges, we introduce a novel and generalizable framework: NeuroBOLT, i.e., Neuro-to-BOLD Transformer, which leverages multi-dimensional representation learning from temporal, spatial, and spectral domains to translate raw EEG data to the corresponding fMRI activity signals across the brain. Our experiments demonstrate that NeuroBOLT effectively reconstructs resting-state fMRI signals from primary sensory, high-level cognitive areas, and deep subcortical brain regions, achieving state-of-the-art accuracy and significantly advancing the integration of these two modalities.
Reconstructing physiological signals from fMRI across the adult lifespan
Aug 26, 2024



Abstract:Interactions between the brain and body are of fundamental importance for human behavior and health. Functional magnetic resonance imaging (fMRI) captures whole-brain activity noninvasively, and modeling how fMRI signals interact with physiological dynamics of the body can provide new insight into brain function and offer potential biomarkers of disease. However, physiological recordings are not always possible to acquire since they require extra equipment and setup, and even when they are, the recorded physiological signals may contain substantial artifacts. To overcome this limitation, machine learning models have been proposed to directly extract features of respiratory and cardiac activity from resting-state fMRI signals. To date, such work has been carried out only in healthy young adults and in a pediatric population, leaving open questions about the efficacy of these approaches on older adults. Here, we propose a novel framework that leverages Transformer-based architectures for reconstructing two key physiological signals - low-frequency respiratory volume (RV) and heart rate (HR) fluctuations - from fMRI data, and test these models on a dataset of individuals aged 36-89 years old. Our framework outperforms previously proposed approaches (attaining median correlations between predicted and measured signals of r ~ .698 for RV and r ~ .618 for HR), indicating the potential of leveraging attention mechanisms to model fMRI-physiological signal relationships. We also evaluate several model training and fine-tuning strategies, and find that incorporating young-adult data during training improves the performance when predicting physiological signals in the aging cohort. Overall, our approach successfully infers key physiological variables directly from fMRI data from individuals across a wide range of the adult lifespan.
Zero-Shot Surgical Tool Segmentation in Monocular Video Using Segment Anything Model 2
Aug 03, 2024Abstract:The Segment Anything Model 2 (SAM 2) is the latest generation foundation model for image and video segmentation. Trained on the expansive Segment Anything Video (SA-V) dataset, which comprises 35.5 million masks across 50.9K videos, SAM 2 advances its predecessor's capabilities by supporting zero-shot segmentation through various prompts (e.g., points, boxes, and masks). Its robust zero-shot performance and efficient memory usage make SAM 2 particularly appealing for surgical tool segmentation in videos, especially given the scarcity of labeled data and the diversity of surgical procedures. In this study, we evaluate the zero-shot video segmentation performance of the SAM 2 model across different types of surgeries, including endoscopy and microscopy. We also assess its performance on videos featuring single and multiple tools of varying lengths to demonstrate SAM 2's applicability and effectiveness in the surgical domain. We found that: 1) SAM 2 demonstrates a strong capability for segmenting various surgical videos; 2) When new tools enter the scene, additional prompts are necessary to maintain segmentation accuracy; and 3) Specific challenges inherent to surgical videos can impact the robustness of SAM 2.
DaReNeRF: Direction-aware Representation for Dynamic Scenes
Mar 04, 2024



Abstract:Addressing the intricate challenge of modeling and re-rendering dynamic scenes, most recent approaches have sought to simplify these complexities using plane-based explicit representations, overcoming the slow training time issues associated with methods like Neural Radiance Fields (NeRF) and implicit representations. However, the straightforward decomposition of 4D dynamic scenes into multiple 2D plane-based representations proves insufficient for re-rendering high-fidelity scenes with complex motions. In response, we present a novel direction-aware representation (DaRe) approach that captures scene dynamics from six different directions. This learned representation undergoes an inverse dual-tree complex wavelet transformation (DTCWT) to recover plane-based information. DaReNeRF computes features for each space-time point by fusing vectors from these recovered planes. Combining DaReNeRF with a tiny MLP for color regression and leveraging volume rendering in training yield state-of-the-art performance in novel view synthesis for complex dynamic scenes. Notably, to address redundancy introduced by the six real and six imaginary direction-aware wavelet coefficients, we introduce a trainable masking approach, mitigating storage issues without significant performance decline. Moreover, DaReNeRF maintains a 2x reduction in training time compared to prior art while delivering superior performance.
Leveraging sinusoidal representation networks to predict fMRI signals from EEG
Nov 06, 2023Abstract:In modern neuroscience, functional magnetic resonance imaging (fMRI) has been a crucial and irreplaceable tool that provides a non-invasive window into the dynamics of whole-brain activity. Nevertheless, fMRI is limited by hemodynamic blurring as well as high cost, immobility, and incompatibility with metal implants. Electroencephalography (EEG) is complementary to fMRI and can directly record the cortical electrical activity at high temporal resolution, but has more limited spatial resolution and is unable to recover information about deep subcortical brain structures. The ability to obtain fMRI information from EEG would enable cost-effective, imaging across a wider set of brain regions. Further, beyond augmenting the capabilities of EEG, cross-modality models would facilitate the interpretation of fMRI signals. However, as both EEG and fMRI are high-dimensional and prone to artifacts, it is currently challenging to model fMRI from EEG. To address this challenge, we propose a novel architecture that can predict fMRI signals directly from multi-channel EEG without explicit feature engineering. Our model achieves this by implementing a Sinusoidal Representation Network (SIREN) to learn frequency information in brain dynamics from EEG, which serves as the input to a subsequent encoder-decoder to effectively reconstruct the fMRI signal from a specific brain region. We evaluate our model using a simultaneous EEG-fMRI dataset with 8 subjects and investigate its potential for predicting subcortical fMRI signals. The present results reveal that our model outperforms a recent state-of-the-art model, and indicates the potential of leveraging periodic activation functions in deep neural networks to model functional neuroimaging data.
SAMSNeRF: Segment Anything Model (SAM) Guides Dynamic Surgical Scene Reconstruction by Neural Radiance Field (NeRF)
Aug 22, 2023Abstract:The accurate reconstruction of surgical scenes from surgical videos is critical for various applications, including intraoperative navigation and image-guided robotic surgery automation. However, previous approaches, mainly relying on depth estimation, have limited effectiveness in reconstructing surgical scenes with moving surgical tools. To address this limitation and provide accurate 3D position prediction for surgical tools in all frames, we propose a novel approach called SAMSNeRF that combines Segment Anything Model (SAM) and Neural Radiance Field (NeRF) techniques. Our approach generates accurate segmentation masks of surgical tools using SAM, which guides the refinement of the dynamic surgical scene reconstruction by NeRF. Our experimental results on public endoscopy surgical videos demonstrate that our approach successfully reconstructs high-fidelity dynamic surgical scenes and accurately reflects the spatial information of surgical tools. Our proposed approach can significantly enhance surgical navigation and automation by providing surgeons with accurate 3D position information of surgical tools during surgery.The source code will be released soon.
AFFIRM: Affinity Fusion-based Framework for Iteratively Random Motion correction of multi-slice fetal brain MRI
May 12, 2022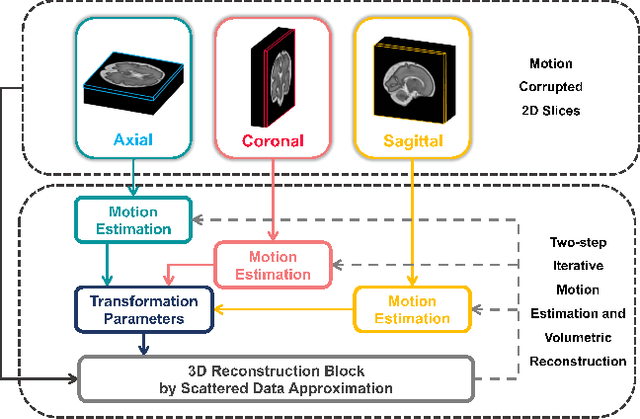
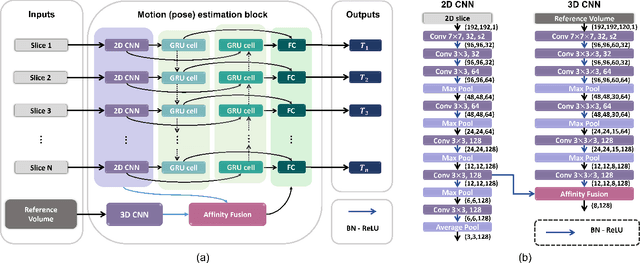
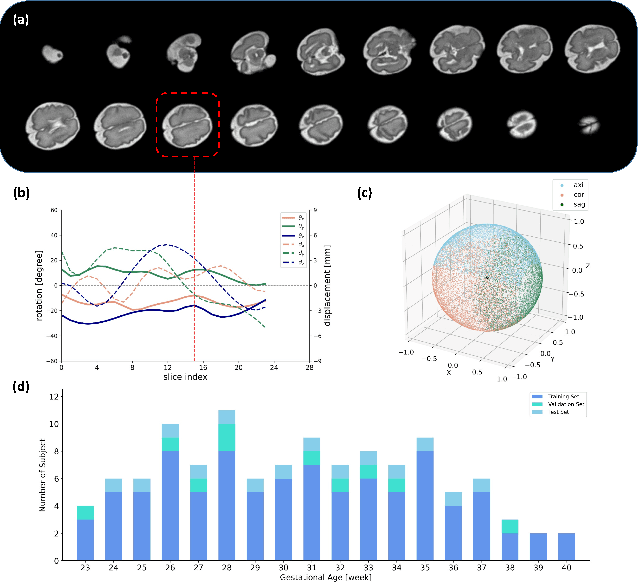
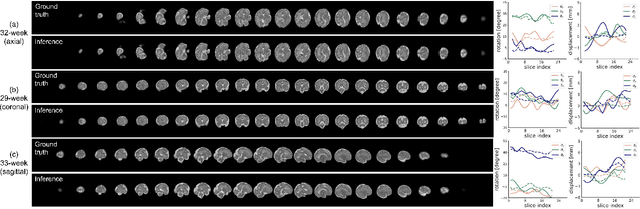
Abstract:Multi-slice magnetic resonance images of the fetal brain are usually contaminated by severe and arbitrary fetal and maternal motion. Hence, stable and robust motion correction is necessary to reconstruct high-resolution 3D fetal brain volume for clinical diagnosis and quantitative analysis. However, the conventional registration-based correction has a limited capture range and is insufficient for detecting relatively large motions. Here, we present a novel Affinity Fusion-based Framework for Iteratively Random Motion (AFFIRM) correction of the multi-slice fetal brain MRI. It learns the sequential motion from multiple stacks of slices and integrates the features between 2D slices and reconstructed 3D volume using affinity fusion, which resembles the iterations between slice-to-volume registration and volumetric reconstruction in the regular pipeline. The method accurately estimates the motion regardless of brain orientations and outperforms other state-of-the-art learning-based methods on the simulated motion-corrupted data, with a 48.4% reduction of mean absolute error for rotation and 61.3% for displacement. We then incorporated AFFIRM into the multi-resolution slice-to-volume registration and tested it on the real-world fetal MRI scans at different gestation stages. The results indicated that adding AFFIRM to the conventional pipeline improved the success rate of fetal brain super-resolution reconstruction from 77.2% to 91.9%.
One Million Scenes for Autonomous Driving: ONCE Dataset
Jun 21, 2021
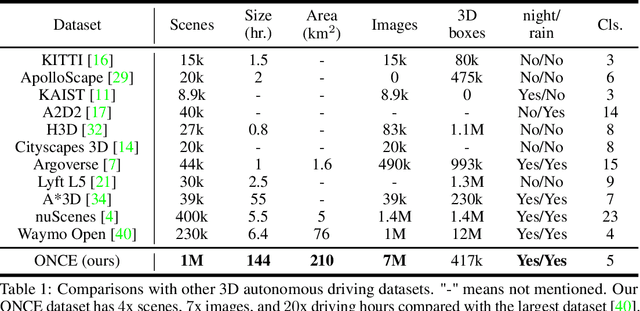
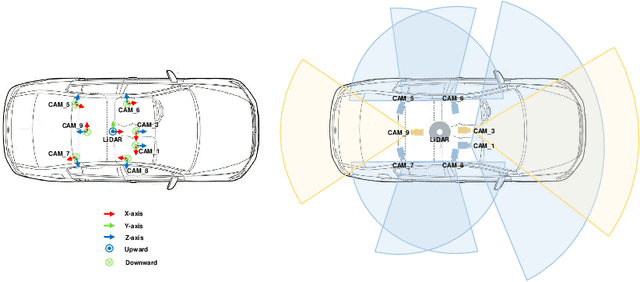
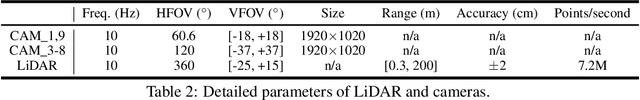
Abstract:Current perception models in autonomous driving have become notorious for greatly relying on a mass of annotated data to cover unseen cases and address the long-tail problem. On the other hand, learning from unlabeled large-scale collected data and incrementally self-training powerful recognition models have received increasing attention and may become the solutions of next-generation industry-level powerful and robust perception models in autonomous driving. However, the research community generally suffered from data inadequacy of those essential real-world scene data, which hampers the future exploration of fully/semi/self-supervised methods for 3D perception. In this paper, we introduce the ONCE (One millioN sCenEs) dataset for 3D object detection in the autonomous driving scenario. The ONCE dataset consists of 1 million LiDAR scenes and 7 million corresponding camera images. The data is selected from 144 driving hours, which is 20x longer than the largest 3D autonomous driving dataset available (e.g. nuScenes and Waymo), and it is collected across a range of different areas, periods and weather conditions. To facilitate future research on exploiting unlabeled data for 3D detection, we additionally provide a benchmark in which we reproduce and evaluate a variety of self-supervised and semi-supervised methods on the ONCE dataset. We conduct extensive analyses on those methods and provide valuable observations on their performance related to the scale of used data. Data, code, and more information are available at https://once-for-auto-driving.github.io/index.html.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge