Guangbin Wang
AtlasSeg: Atlas Prior Guided Dual-U-Net for Cortical Segmentation in Fetal Brain MRI
Nov 05, 2024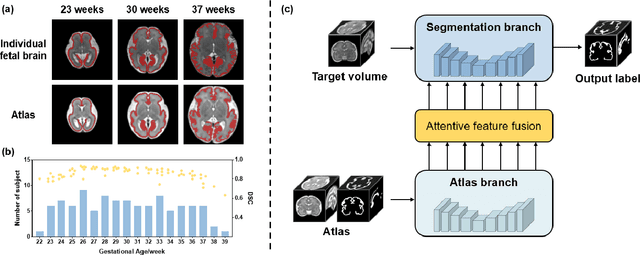
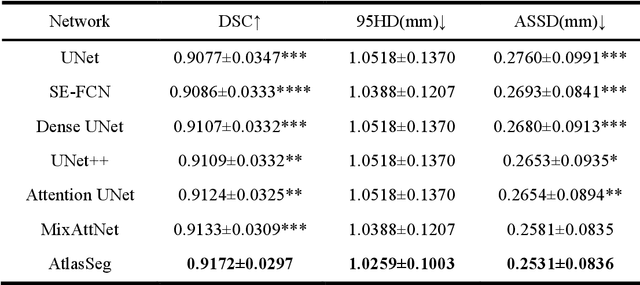
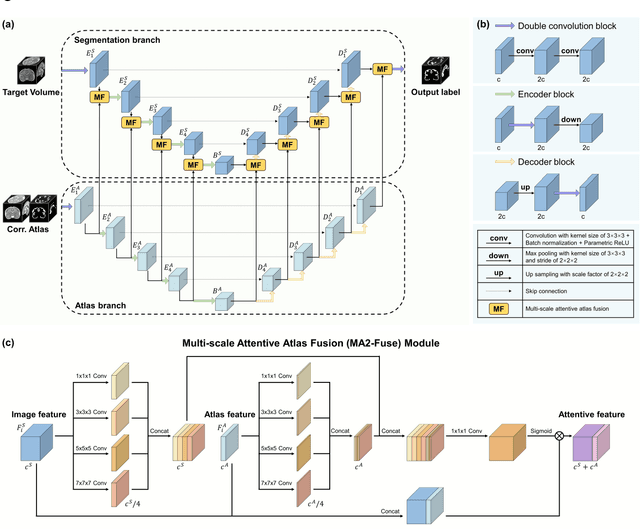

Abstract:Accurate tissue segmentation in fetal brain MRI remains challenging due to the dynamically changing anatomical anatomy and contrast during fetal development. To enhance segmentation accuracy throughout gestation, we introduced AtlasSeg, a dual-U-shape convolution network incorporating gestational age (GA) specific information as guidance. By providing a publicly available fetal brain atlas with segmentation label at the corresponding GA, AtlasSeg effectively extracted the contextual features of age-specific patterns in atlas branch and generated tissue segmentation in segmentation branch. Multi-scale attentive atlas feature fusions were constructed in all stages during encoding and decoding, giving rise to a dual-U-shape network to assist feature flow and information interactions between two branches. AtlasSeg outperformed six well-known segmentation networks in both our internal fetal brain MRI dataset and the external FeTA dataset. Ablation experiments demonstrate the efficiency of atlas guidance and the attention mechanism. The proposed AtlasSeg demonstrated superior segmentation performance against other convolution networks with higher segmentation accuracy, and may facilitate fetal brain MRI analysis in large-scale fetal brain studies.
A Motion Assessment Method for Reference Stack Selection in Fetal Brain MRI Reconstruction Based on Tensor Rank Approximation
Jun 30, 2023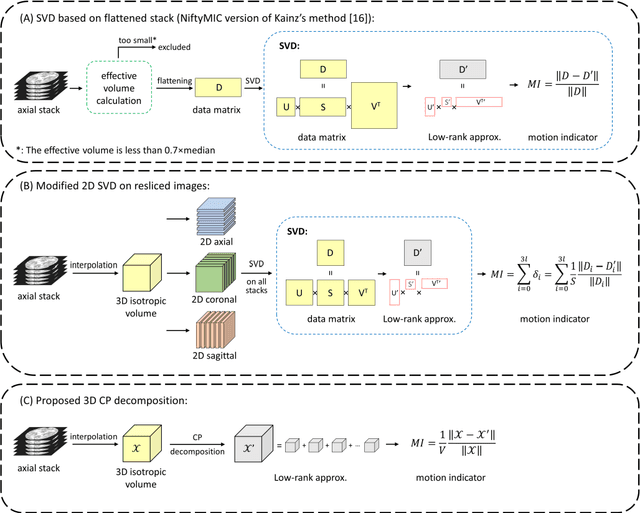
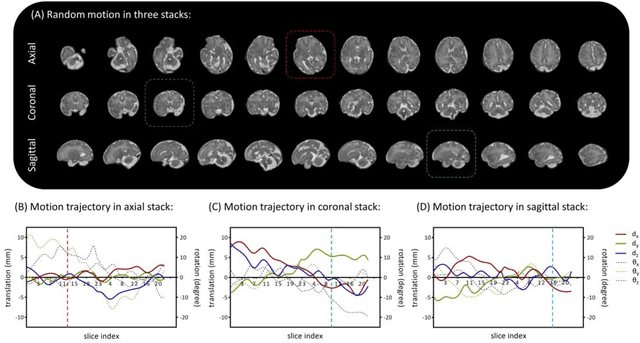
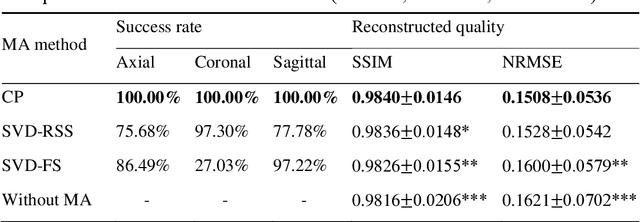
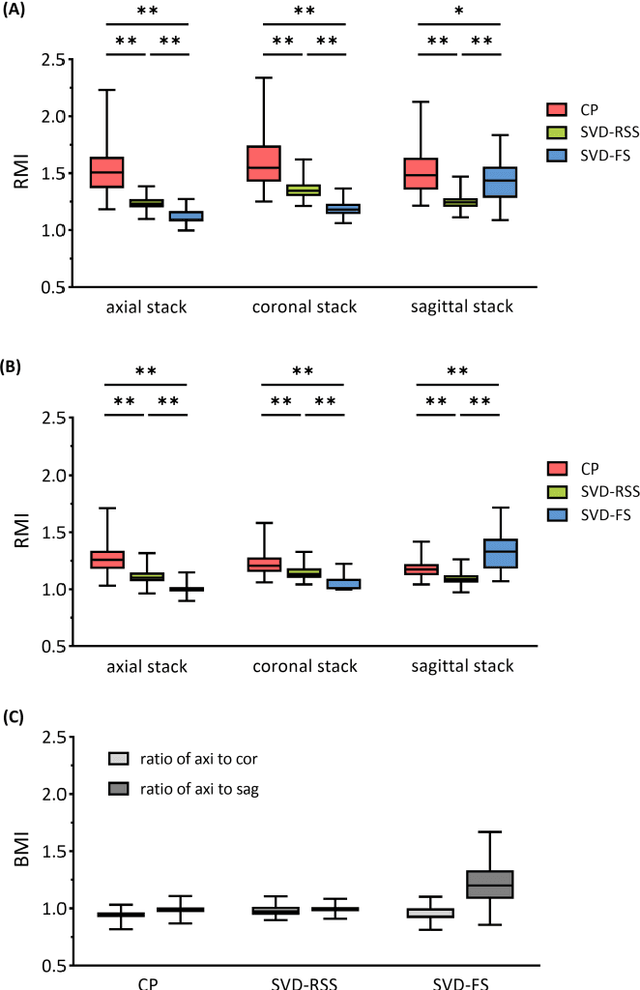
Abstract:Purpose: Slice-to-volume registration and super-resolution reconstruction (SVR-SRR) is commonly used to generate 3D volumes of the fetal brain from 2D stacks of slices acquired in multiple orientations. A critical initial step in this pipeline is to select one stack with the minimum motion as a reference for registration. An accurate and unbiased motion assessment (MA) is thus crucial for successful selection. Methods: We presented a MA method that determines the minimum motion stack based on 3D low-rank approximation using CANDECOMP/PARAFAC (CP) decomposition. Compared to the current 2D singular value decomposition (SVD) based method that requires flattening stacks into matrices to obtain ranks, in which the spatial information is lost, the CP-based method can factorize 3D stack into low-rank and sparse components in a computationally efficient manner. The difference between the original stack and its low-rank approximation was proposed as the motion indicator. Results: Compared to SVD-based methods, our proposed CP-based MA demonstrated higher sensitivity in detecting small motion with a lower baseline bias. Experiments on randomly simulated motion illustrated that the proposed CP method achieved a higher success rate of 95.45% in identifying the minimum motion stack, compared to SVD-based method with a success rate of 58.18%. We further demonstrated that combining CP-based MA with existing SRR-SVR pipeline significantly improved 3D volume reconstruction. Conclusion: The proposed CP-based MA method showed superior performance compared to SVD-based methods with higher sensitivity to motion, success rate, and lower baseline bias, and can be used as a prior step to improve fetal brain reconstruction.
A microstructure estimation Transformer inspired by sparse representation for diffusion MRI
May 13, 2022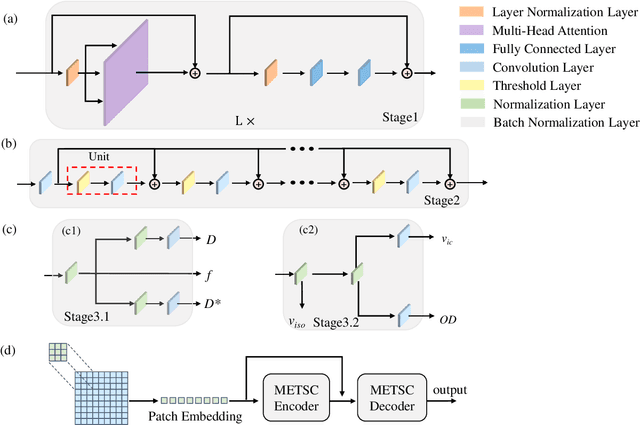
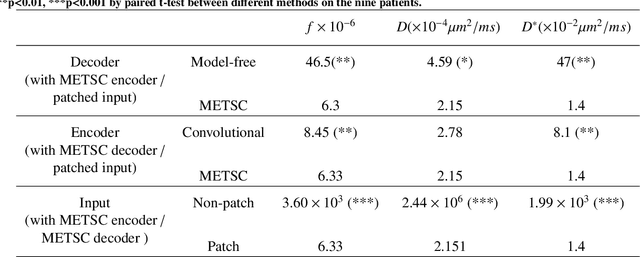
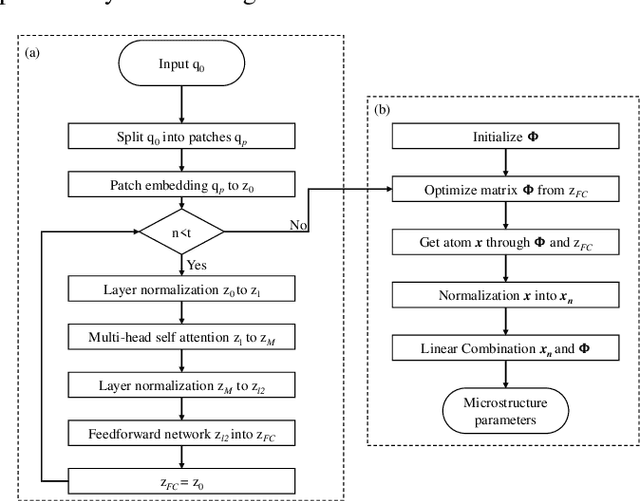
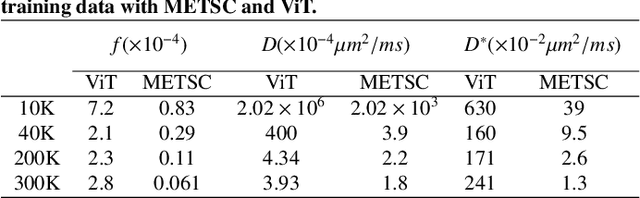
Abstract:Diffusion magnetic resonance imaging (dMRI) is an important tool in characterizing tissue microstructure based on biophysical models, which are complex and highly non-linear. Resolving microstructures with optimization techniques is prone to estimation errors and requires dense sampling in the q-space. Deep learning based approaches have been proposed to overcome these limitations. Motivated by the superior performance of the Transformer, in this work, we present a learning-based framework based on Transformer, namely, a Microstructure Estimation Transformer with Sparse Coding (METSC) for dMRI-based microstructure estimation with downsampled q-space data. To take advantage of the Transformer while addressing its limitation in large training data requirements, we explicitly introduce an inductive bias - model bias into the Transformer using a sparse coding technique to facilitate the training process. Thus, the METSC is composed with three stages, an embedding stage, a sparse representation stage, and a mapping stage. The embedding stage is a Transformer-based structure that encodes the signal to ensure the voxel is represented effectively. In the sparse representation stage, a dictionary is constructed by solving a sparse reconstruction problem that unfolds the Iterative Hard Thresholding (IHT) process. The mapping stage is essentially a decoder that computes the microstructural parameters from the output of the second stage, based on the weighted sum of normalized dictionary coefficients where the weights are also learned. We tested our framework on two dMRI models with downsampled q-space data, including the intravoxel incoherent motion (IVIM) model and the neurite orientation dispersion and density imaging (NODDI) model. The proposed method achieved up to 11.25 folds of acceleration in scan time and outperformed the other state-of-the-art learning-based methods.
AFFIRM: Affinity Fusion-based Framework for Iteratively Random Motion correction of multi-slice fetal brain MRI
May 12, 2022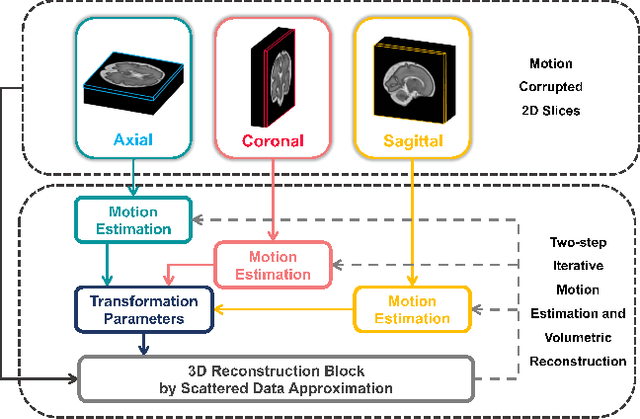
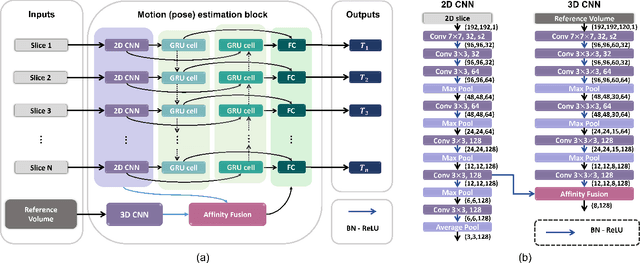
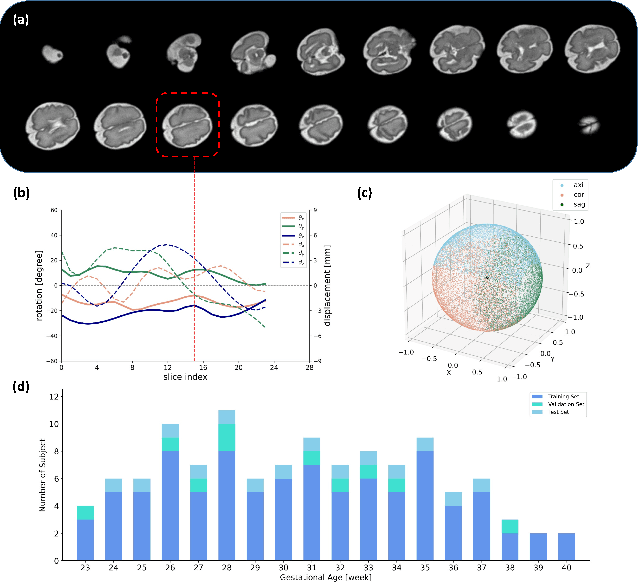
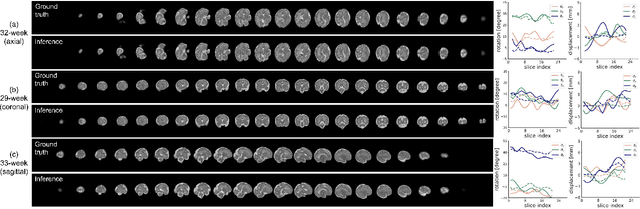
Abstract:Multi-slice magnetic resonance images of the fetal brain are usually contaminated by severe and arbitrary fetal and maternal motion. Hence, stable and robust motion correction is necessary to reconstruct high-resolution 3D fetal brain volume for clinical diagnosis and quantitative analysis. However, the conventional registration-based correction has a limited capture range and is insufficient for detecting relatively large motions. Here, we present a novel Affinity Fusion-based Framework for Iteratively Random Motion (AFFIRM) correction of the multi-slice fetal brain MRI. It learns the sequential motion from multiple stacks of slices and integrates the features between 2D slices and reconstructed 3D volume using affinity fusion, which resembles the iterations between slice-to-volume registration and volumetric reconstruction in the regular pipeline. The method accurately estimates the motion regardless of brain orientations and outperforms other state-of-the-art learning-based methods on the simulated motion-corrupted data, with a 48.4% reduction of mean absolute error for rotation and 61.3% for displacement. We then incorporated AFFIRM into the multi-resolution slice-to-volume registration and tested it on the real-world fetal MRI scans at different gestation stages. The results indicated that adding AFFIRM to the conventional pipeline improved the success rate of fetal brain super-resolution reconstruction from 77.2% to 91.9%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge