A microstructure estimation Transformer inspired by sparse representation for diffusion MRI
Paper and Code
May 13, 2022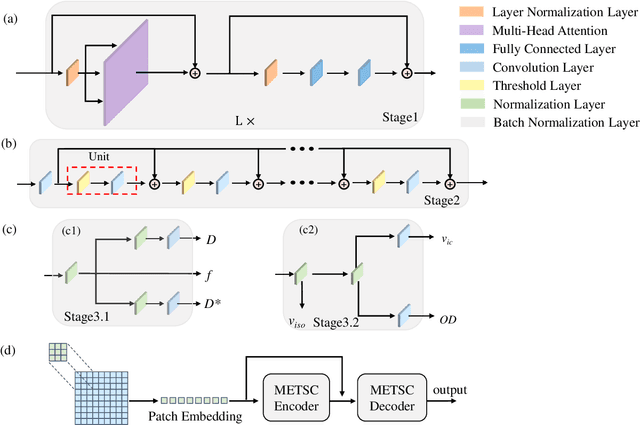
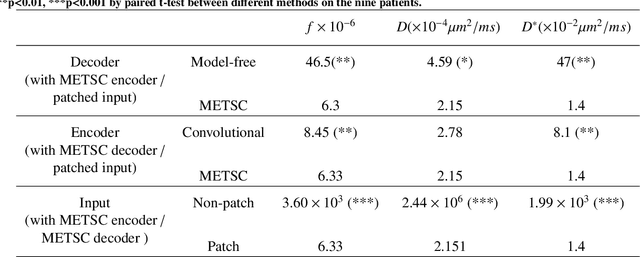
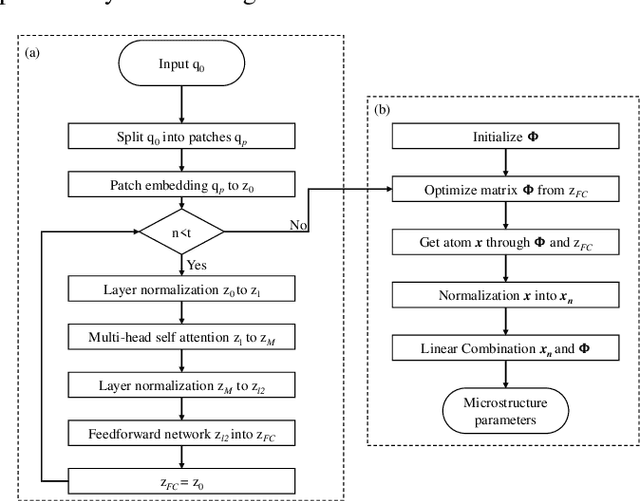
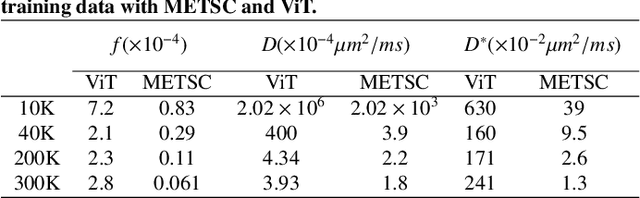
Diffusion magnetic resonance imaging (dMRI) is an important tool in characterizing tissue microstructure based on biophysical models, which are complex and highly non-linear. Resolving microstructures with optimization techniques is prone to estimation errors and requires dense sampling in the q-space. Deep learning based approaches have been proposed to overcome these limitations. Motivated by the superior performance of the Transformer, in this work, we present a learning-based framework based on Transformer, namely, a Microstructure Estimation Transformer with Sparse Coding (METSC) for dMRI-based microstructure estimation with downsampled q-space data. To take advantage of the Transformer while addressing its limitation in large training data requirements, we explicitly introduce an inductive bias - model bias into the Transformer using a sparse coding technique to facilitate the training process. Thus, the METSC is composed with three stages, an embedding stage, a sparse representation stage, and a mapping stage. The embedding stage is a Transformer-based structure that encodes the signal to ensure the voxel is represented effectively. In the sparse representation stage, a dictionary is constructed by solving a sparse reconstruction problem that unfolds the Iterative Hard Thresholding (IHT) process. The mapping stage is essentially a decoder that computes the microstructural parameters from the output of the second stage, based on the weighted sum of normalized dictionary coefficients where the weights are also learned. We tested our framework on two dMRI models with downsampled q-space data, including the intravoxel incoherent motion (IVIM) model and the neurite orientation dispersion and density imaging (NODDI) model. The proposed method achieved up to 11.25 folds of acceleration in scan time and outperformed the other state-of-the-art learning-based methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge