A Motion Assessment Method for Reference Stack Selection in Fetal Brain MRI Reconstruction Based on Tensor Rank Approximation
Paper and Code
Jun 30, 2023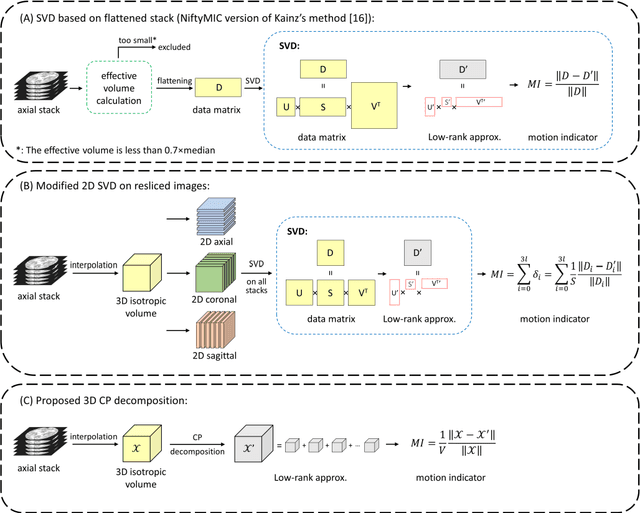
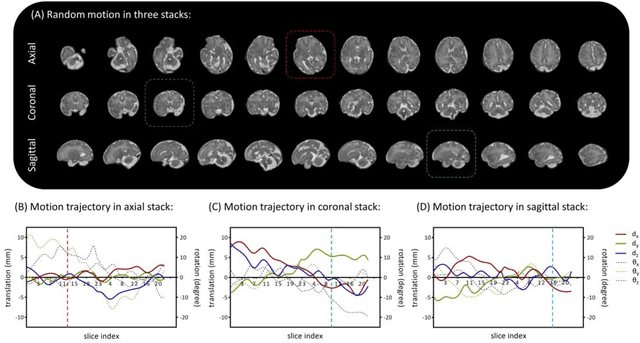
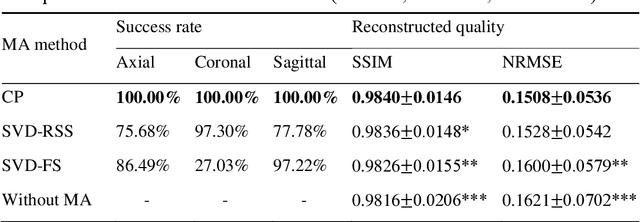
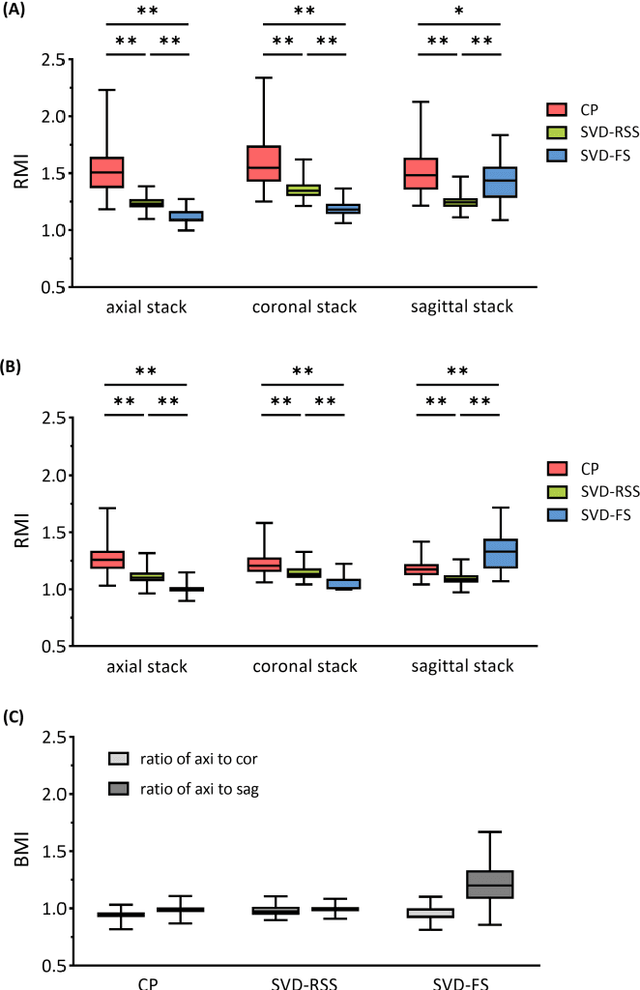
Purpose: Slice-to-volume registration and super-resolution reconstruction (SVR-SRR) is commonly used to generate 3D volumes of the fetal brain from 2D stacks of slices acquired in multiple orientations. A critical initial step in this pipeline is to select one stack with the minimum motion as a reference for registration. An accurate and unbiased motion assessment (MA) is thus crucial for successful selection. Methods: We presented a MA method that determines the minimum motion stack based on 3D low-rank approximation using CANDECOMP/PARAFAC (CP) decomposition. Compared to the current 2D singular value decomposition (SVD) based method that requires flattening stacks into matrices to obtain ranks, in which the spatial information is lost, the CP-based method can factorize 3D stack into low-rank and sparse components in a computationally efficient manner. The difference between the original stack and its low-rank approximation was proposed as the motion indicator. Results: Compared to SVD-based methods, our proposed CP-based MA demonstrated higher sensitivity in detecting small motion with a lower baseline bias. Experiments on randomly simulated motion illustrated that the proposed CP method achieved a higher success rate of 95.45% in identifying the minimum motion stack, compared to SVD-based method with a success rate of 58.18%. We further demonstrated that combining CP-based MA with existing SRR-SVR pipeline significantly improved 3D volume reconstruction. Conclusion: The proposed CP-based MA method showed superior performance compared to SVD-based methods with higher sensitivity to motion, success rate, and lower baseline bias, and can be used as a prior step to improve fetal brain reconstruction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge