Tengfei Cao
Toward Stable Semi-Supervised Remote Sensing Segmentation via Co-Guidance and Co-Fusion
Dec 28, 2025Abstract:Semi-supervised remote sensing (RS) image semantic segmentation offers a promising solution to alleviate the burden of exhaustive annotation, yet it fundamentally struggles with pseudo-label drift, a phenomenon where confirmation bias leads to the accumulation of errors during training. In this work, we propose Co2S, a stable semi-supervised RS segmentation framework that synergistically fuses priors from vision-language models and self-supervised models. Specifically, we construct a heterogeneous dual-student architecture comprising two distinct ViT-based vision foundation models initialized with pretrained CLIP and DINOv3 to mitigate error accumulation and pseudo-label drift. To effectively incorporate these distinct priors, an explicit-implicit semantic co-guidance mechanism is introduced that utilizes text embeddings and learnable queries to provide explicit and implicit class-level guidance, respectively, thereby jointly enhancing semantic consistency. Furthermore, a global-local feature collaborative fusion strategy is developed to effectively fuse the global contextual information captured by CLIP with the local details produced by DINOv3, enabling the model to generate highly precise segmentation results. Extensive experiments on six popular datasets demonstrate the superiority of the proposed method, which consistently achieves leading performance across various partition protocols and diverse scenarios. Project page is available at https://xavierjiezou.github.io/Co2S/.
Knowledge Transfer and Domain Adaptation for Fine-Grained Remote Sensing Image Segmentation
Dec 09, 2024



Abstract:Fine-grained remote sensing image segmentation is essential for accurately identifying detailed objects in remote sensing images. Recently, vision transformer models (VTM) pretrained on large-scale datasets have shown strong zero-shot generalization, indicating that they have learned the general knowledge of object understanding. We introduce a novel end-to-end learning paradigm combining knowledge guidance with domain refinement to enhance performance. We present two key components: the Feature Alignment Module (FAM) and the Feature Modulation Module (FMM). FAM aligns features from a CNN-based backbone with those from the pretrained VTM's encoder using channel transformation and spatial interpolation, and transfers knowledge via KL divergence and L2 normalization constraint. FMM further adapts the knowledge to the specific domain to address domain shift. We also introduce a fine-grained grass segmentation dataset and demonstrate, through experiments on two datasets, that our method achieves a significant improvement of 2.57 mIoU on the grass dataset and 3.73 mIoU on the cloud dataset. The results highlight the potential of combining knowledge transfer and domain adaptation to overcome domain-related challenges and data limitations. The project page is available at https://xavierjiezou.github.io/KTDA/.
Hunyuan-Large: An Open-Source MoE Model with 52 Billion Activated Parameters by Tencent
Nov 05, 2024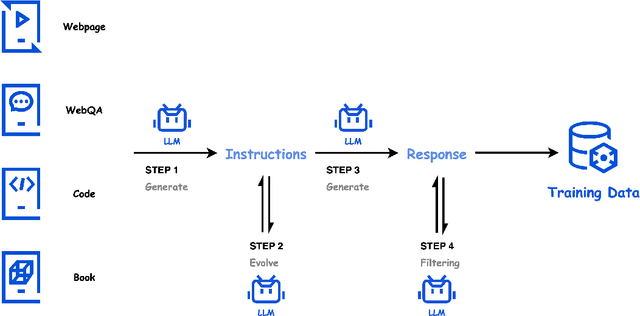
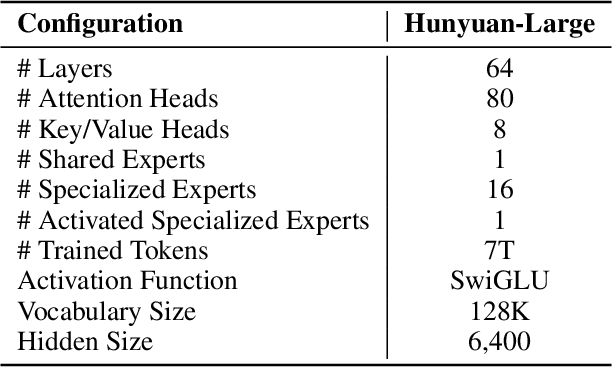
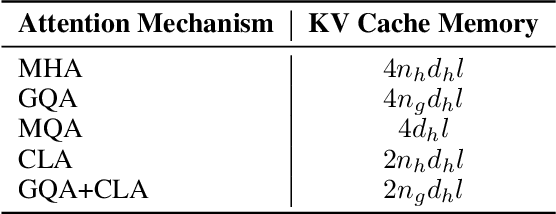
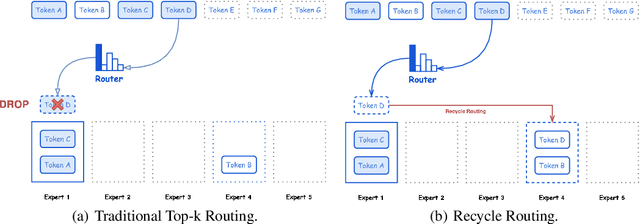
Abstract:In this paper, we introduce Hunyuan-Large, which is currently the largest open-source Transformer-based mixture of experts model, with a total of 389 billion parameters and 52 billion activation parameters, capable of handling up to 256K tokens. We conduct a thorough evaluation of Hunyuan-Large's superior performance across various benchmarks including language understanding and generation, logical reasoning, mathematical problem-solving, coding, long-context, and aggregated tasks, where it outperforms LLama3.1-70B and exhibits comparable performance when compared to the significantly larger LLama3.1-405B model. Key practice of Hunyuan-Large include large-scale synthetic data that is orders larger than in previous literature, a mixed expert routing strategy, a key-value cache compression technique, and an expert-specific learning rate strategy. Additionally, we also investigate the scaling laws and learning rate schedule of mixture of experts models, providing valuable insights and guidances for future model development and optimization. The code and checkpoints of Hunyuan-Large are released to facilitate future innovations and applications. Codes: https://github.com/Tencent/Hunyuan-Large Models: https://huggingface.co/tencent/Tencent-Hunyuan-Large
Capture Agent Free Biosensing using Porous Silicon Arrays and Machine Learning
Jan 22, 2022



Abstract:Biosensors are an essential tool for medical diagnostics, environmental monitoring and food safety. Typically, biosensors are designed to detect specific analytes through functionalization with the appropriate capture agents. However, the use of capture agents limits the number of analytes that can be simultaneously detected and reduces the robustness of the biosensor. In this work, we report a versatile, capture agent free biosensor platform based on an array of porous silicon (PSi) thin films, which has the potential to robustly detect a wide variety of analytes based on their physical and chemical properties in the nanoscale porous media. The ability of this system to reproducibly classify, quantify, and discriminate three proteins is demonstrated to concentrations down to at least 0.02g/L (between 300nM and 450nM) by utilizing PSi array elements with a unique combination of pore size and buffer pH, employing linear discriminant analysis for dimensionality reduction, and using support vector machines as a classifier. This approach represents a significant step towards a low cost, simple and robust biosensor platform that is able to detect a vast range of biomolecules.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge