Lee E. Wheless
Segment Anything Model (SAM) for Digital Pathology: Assess Zero-shot Segmentation on Whole Slide Imaging
Apr 09, 2023Abstract:The segment anything model (SAM) was released as a foundation model for image segmentation. The promptable segmentation model was trained by over 1 billion masks on 11M licensed and privacy-respecting images. The model supports zero-shot image segmentation with various segmentation prompts (e.g., points, boxes, masks). It makes the SAM attractive for medical image analysis, especially for digital pathology where the training data are rare. In this study, we evaluate the zero-shot segmentation performance of SAM model on representative segmentation tasks on whole slide imaging (WSI), including (1) tumor segmentation, (2) non-tumor tissue segmentation, (3) cell nuclei segmentation. Core Results: The results suggest that the zero-shot SAM model achieves remarkable segmentation performance for large connected objects. However, it does not consistently achieve satisfying performance for dense instance object segmentation, even with 20 prompts (clicks/boxes) on each image. We also summarized the identified limitations for digital pathology: (1) image resolution, (2) multiple scales, (3) prompt selection, and (4) model fine-tuning. In the future, the few-shot fine-tuning with images from downstream pathological segmentation tasks might help the model to achieve better performance in dense object segmentation.
SimTriplet: Simple Triplet Representation Learning with a Single GPU
Mar 09, 2021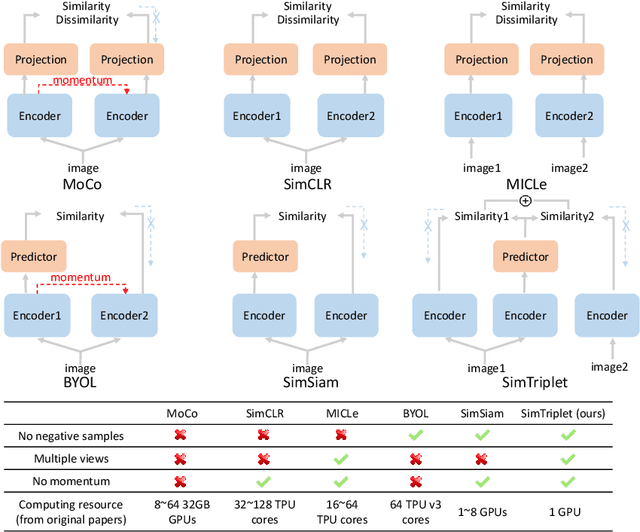
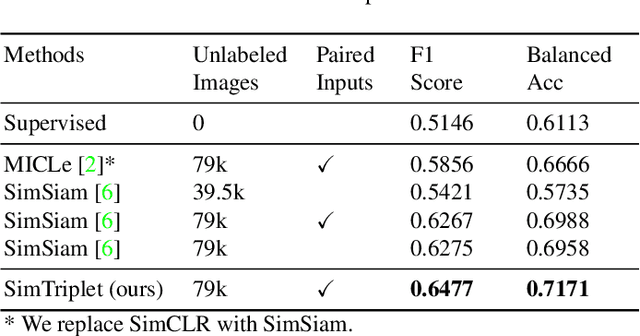
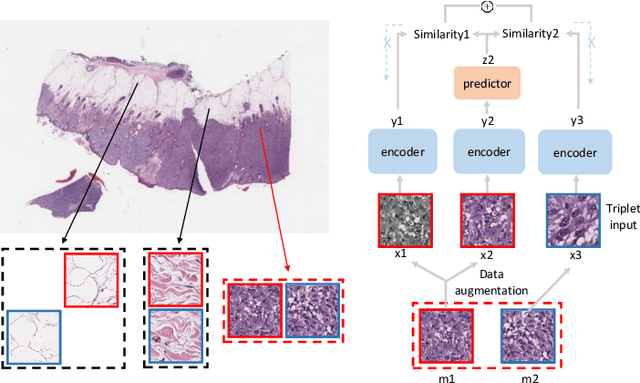
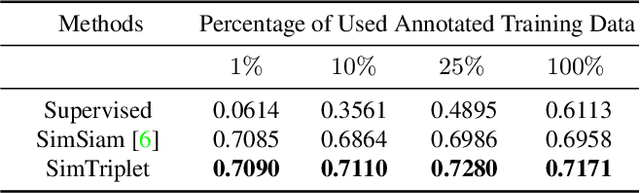
Abstract:Contrastive learning is a key technique of modern self-supervised learning. The broader accessibility of earlier approaches is hindered by the need of heavy computational resources (e.g., at least 8 GPUs or 32 TPU cores), which accommodate for large-scale negative samples or momentum. The more recent SimSiam approach addresses such key limitations via stop-gradient without momentum encoders. In medical image analysis, multiple instances can be achieved from the same patient or tissue. Inspired by these advances, we propose a simple triplet representation learning (SimTriplet) approach on pathological images. The contribution of the paper is three-fold: (1) The proposed SimTriplet method takes advantage of the multi-view nature of medical images beyond self-augmentation; (2) The method maximizes both intra-sample and inter-sample similarities via triplets from positive pairs, without using negative samples; and (3) The recent mix precision training is employed to advance the training by only using a single GPU with 16GB memory. By learning from 79,000 unlabeled pathological patch images, SimTriplet achieved 10.58% better performance compared with supervised learning. It also achieved 2.13% better performance compared with SimSiam. Our proposed SimTriplet can achieve decent performance using only 1% labeled data. The code and data are available at https://github.com/hrlblab/SimTriple.
BEDS: Bagging ensemble deep segmentation for nucleus segmentation with testing stage stain augmentation
Feb 17, 2021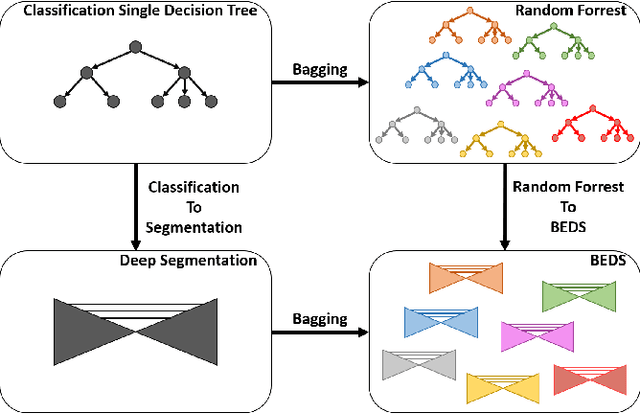
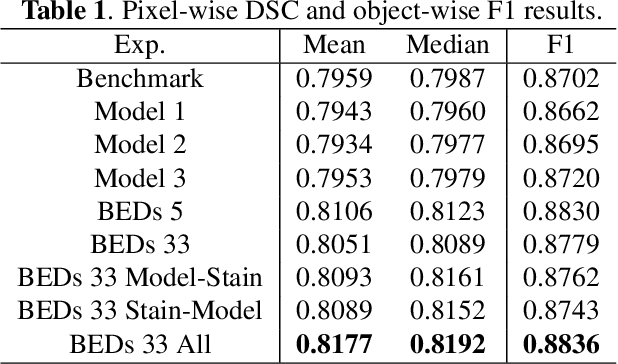
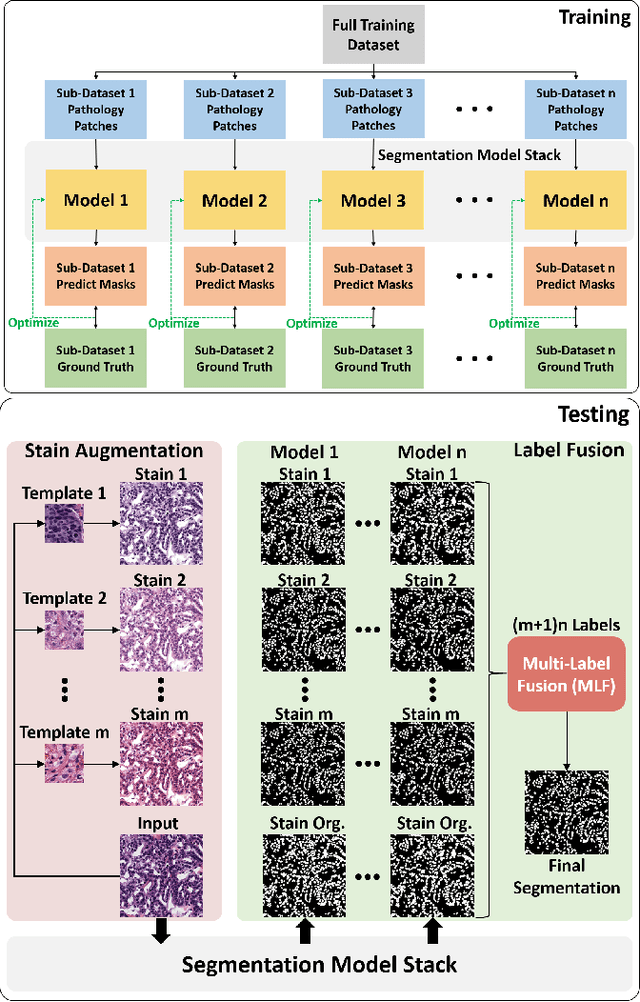

Abstract:Reducing outcome variance is an essential task in deep learning based medical image analysis. Bootstrap aggregating, also known as bagging, is a canonical ensemble algorithm for aggregating weak learners to become a strong learner. Random forest is one of the most powerful machine learning algorithms before deep learning era, whose superior performance is driven by fitting bagged decision trees (weak learners). Inspired by the random forest technique, we propose a simple bagging ensemble deep segmentation (BEDs) method to train multiple U-Nets with partial training data to segment dense nuclei on pathological images. The contributions of this study are three-fold: (1) developing a self-ensemble learning framework for nucleus segmentation; (2) aggregating testing stage augmentation with self-ensemble learning; and (3) elucidating the idea that self-ensemble and testing stage stain augmentation are complementary strategies for a superior segmentation performance. Implementation Detail: https://github.com/xingli1102/BEDs.
WearMask: Fast In-browser Face Mask Detection with Serverless Edge Computing for COVID-19
Jan 04, 2021



Abstract:The COVID-19 epidemic has been a significant healthcare challenge in the United States. According to the Centers for Disease Control and Prevention (CDC), COVID-19 infection is transmitted predominately by respiratory droplets generated when people breathe, talk, cough, or sneeze. Wearing a mask is the primary, effective, and convenient method of blocking 80% of all respiratory infections. Therefore, many face mask detection and monitoring systems have been developed to provide effective supervision for hospitals, airports, publication transportation, sports venues, and retail locations. However, the current commercial face mask detection systems are typically bundled with specific software or hardware, impeding public accessibility. In this paper, we propose an in-browser serverless edge-computing based face mask detection solution, called Web-based efficient AI recognition of masks (WearMask), which can be deployed on any common devices (e.g., cell phones, tablets, computers) that have internet connections using web browsers, without installing any software. The serverless edge-computing design minimizes the extra hardware costs (e.g., specific devices or cloud computing servers). The contribution of the proposed method is to provide a holistic edge-computing framework of integrating (1) deep learning models (YOLO), (2) high-performance neural network inference computing framework (NCNN), and (3) a stack-based virtual machine (WebAssembly). For end-users, our web-based solution has advantages of (1) serverless edge-computing design with minimal device limitation and privacy risk, (2) installation free deployment, (3) low computing requirements, and (4) high detection speed. Our WearMask application has been launched with public access at facemask-detection.com.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge