Yuwei Yang
Effective Training Data Synthesis for Improving MLLM Chart Understanding
Aug 08, 2025Abstract:Being able to effectively read scientific plots, or chart understanding, is a central part toward building effective agents for science. However, existing multimodal large language models (MLLMs), especially open-source ones, are still falling behind with a typical success rate of 30%-50% on challenging benchmarks. Previous studies on fine-tuning MLLMs with synthetic charts are often restricted by their inadequate similarity to the real charts, which could compromise model training and performance on complex real-world charts. In this study, we show that modularizing chart generation and diversifying visual details improves chart understanding capabilities. In particular, we design a five-step data synthesis pipeline, where we separate data and function creation for single plot generation, condition the generation of later subplots on earlier ones for multi-subplot figures, visually diversify the generated figures, filter out low quality data, and finally generate the question-answer (QA) pairs with GPT-4o. This approach allows us to streamline the generation of fine-tuning datasets and introduce the effective chart dataset (ECD), which contains 10k+ chart images and 300k+ QA pairs, covering 25 topics and featuring 250+ chart type combinations with high visual complexity. We show that ECD consistently improves the performance of various MLLMs on a range of real-world and synthetic test sets. Code, data and models are available at: https://github.com/yuweiyang-anu/ECD.
Exploring Gender Bias in Alzheimer's Disease Detection: Insights from Mandarin and Greek Speech Perception
Jul 16, 2025
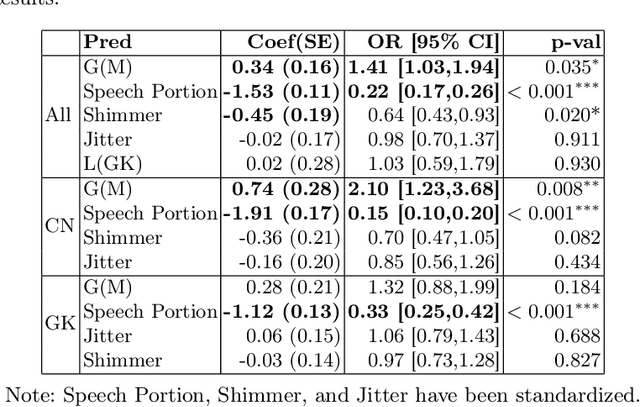
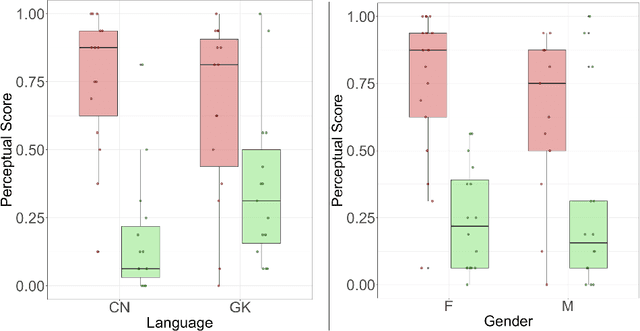
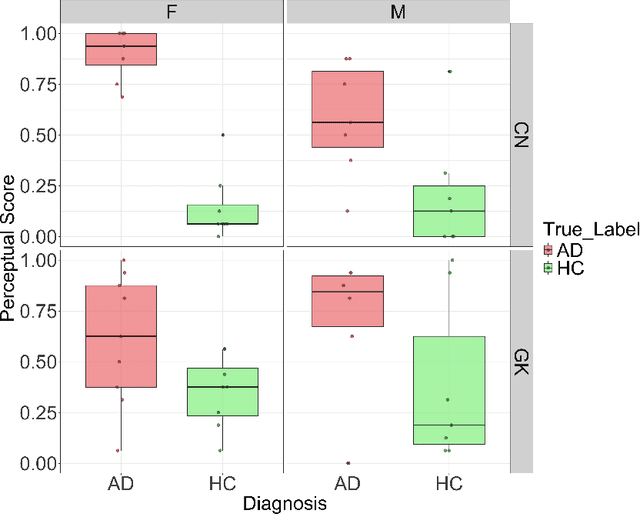
Abstract:Gender bias has been widely observed in speech perception tasks, influenced by the fundamental voicing differences between genders. This study reveals a gender bias in the perception of Alzheimer's Disease (AD) speech. In a perception experiment involving 16 Chinese listeners evaluating both Chinese and Greek speech, we identified that male speech was more frequently identified as AD, with this bias being particularly pronounced in Chinese speech. Acoustic analysis showed that shimmer values in male speech were significantly associated with AD perception, while speech portion exhibited a significant negative correlation with AD identification. Although language did not have a significant impact on AD perception, our findings underscore the critical role of gender bias in AD speech perception. This work highlights the necessity of addressing gender bias when developing AD detection models and calls for further research to validate model performance across different linguistic contexts.
MMMG: A Massive, Multidisciplinary, Multi-Tier Generation Benchmark for Text-to-Image Reasoning
Jun 12, 2025Abstract:In this paper, we introduce knowledge image generation as a new task, alongside the Massive Multi-Discipline Multi-Tier Knowledge-Image Generation Benchmark (MMMG) to probe the reasoning capability of image generation models. Knowledge images have been central to human civilization and to the mechanisms of human learning--a fact underscored by dual-coding theory and the picture-superiority effect. Generating such images is challenging, demanding multimodal reasoning that fuses world knowledge with pixel-level grounding into clear explanatory visuals. To enable comprehensive evaluation, MMMG offers 4,456 expert-validated (knowledge) image-prompt pairs spanning 10 disciplines, 6 educational levels, and diverse knowledge formats such as charts, diagrams, and mind maps. To eliminate confounding complexity during evaluation, we adopt a unified Knowledge Graph (KG) representation. Each KG explicitly delineates a target image's core entities and their dependencies. We further introduce MMMG-Score to evaluate generated knowledge images. This metric combines factual fidelity, measured by graph-edit distance between KGs, with visual clarity assessment. Comprehensive evaluations of 16 state-of-the-art text-to-image generation models expose serious reasoning deficits--low entity fidelity, weak relations, and clutter--with GPT-4o achieving an MMMG-Score of only 50.20, underscoring the benchmark's difficulty. To spur further progress, we release FLUX-Reason (MMMG-Score of 34.45), an effective and open baseline that combines a reasoning LLM with diffusion models and is trained on 16,000 curated knowledge image-prompt pairs.
Training-free Dense-Aligned Diffusion Guidance for Modular Conditional Image Synthesis
Apr 03, 2025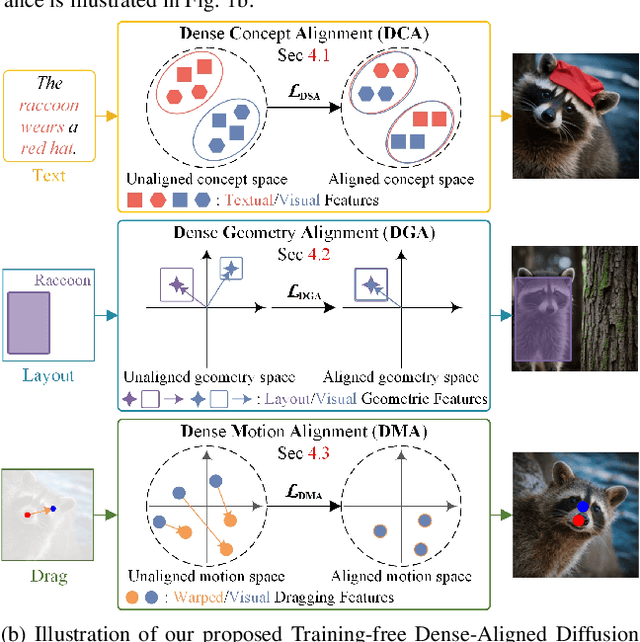
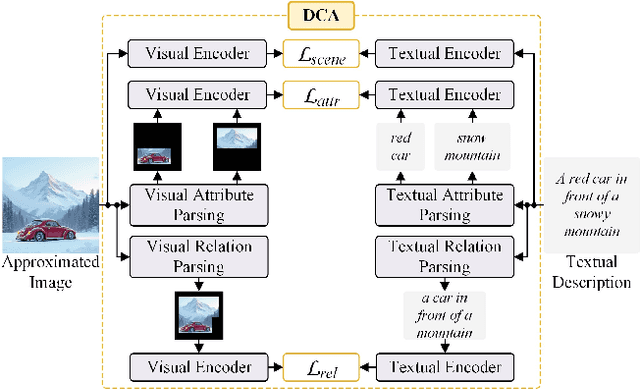
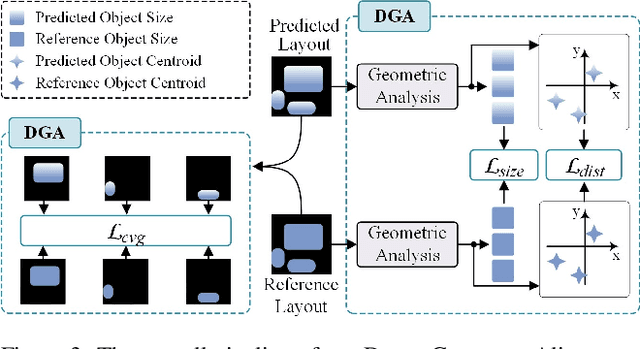
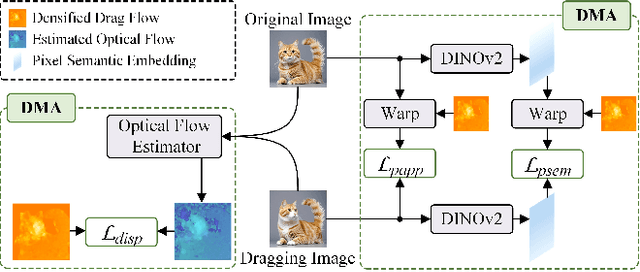
Abstract:Conditional image synthesis is a crucial task with broad applications, such as artistic creation and virtual reality. However, current generative methods are often task-oriented with a narrow scope, handling a restricted condition with constrained applicability. In this paper, we propose a novel approach that treats conditional image synthesis as the modular combination of diverse fundamental condition units. Specifically, we divide conditions into three primary units: text, layout, and drag. To enable effective control over these conditions, we design a dedicated alignment module for each. For the text condition, we introduce a Dense Concept Alignment (DCA) module, which achieves dense visual-text alignment by drawing on diverse textual concepts. For the layout condition, we propose a Dense Geometry Alignment (DGA) module to enforce comprehensive geometric constraints that preserve the spatial configuration. For the drag condition, we introduce a Dense Motion Alignment (DMA) module to apply multi-level motion regularization, ensuring that each pixel follows its desired trajectory without visual artifacts. By flexibly inserting and combining these alignment modules, our framework enhances the model's adaptability to diverse conditional generation tasks and greatly expands its application range. Extensive experiments demonstrate the superior performance of our framework across a variety of conditions, including textual description, segmentation mask (bounding box), drag manipulation, and their combinations. Code is available at https://github.com/ZixuanWang0525/DADG.
Decomposed Direct Preference Optimization for Structure-Based Drug Design
Jul 19, 2024Abstract:Diffusion models have achieved promising results for Structure-Based Drug Design (SBDD). Nevertheless, high-quality protein subpocket and ligand data are relatively scarce, which hinders the models' generation capabilities. Recently, Direct Preference Optimization (DPO) has emerged as a pivotal tool for the alignment of generative models such as large language models and diffusion models, providing greater flexibility and accuracy by directly aligning model outputs with human preferences. Building on this advancement, we introduce DPO to SBDD in this paper. We tailor diffusion models to pharmaceutical needs by aligning them with elaborately designed chemical score functions. We propose a new structure-based molecular optimization method called DecompDPO, which decomposes the molecule into arms and scaffolds and performs preference optimization at both local substructure and global molecule levels, allowing for more precise control with fine-grained preferences. Notably, DecompDPO can be effectively used for two main purposes: (1) fine-tuning pretrained diffusion models for molecule generation across various protein families, and (2) molecular optimization given a specific protein subpocket after generation. Extensive experiments on the CrossDocked2020 benchmark show that DecompDPO significantly improves model performance in both molecule generation and optimization, with up to 100% Median High Affinity and a 54.9% Success Rate.
Structure-Based Drug Design via 3D Molecular Generative Pre-training and Sampling
Feb 22, 2024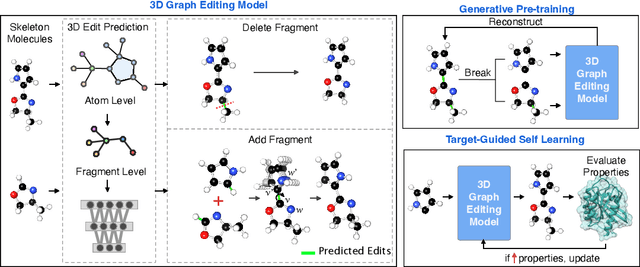
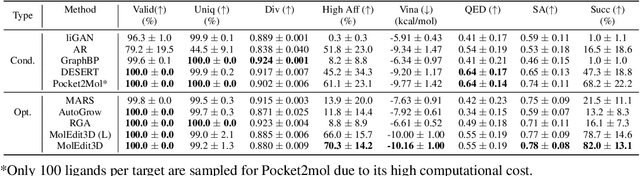
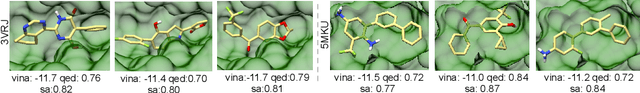
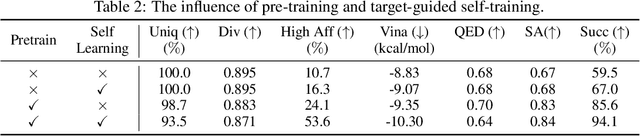
Abstract:Structure-based drug design aims at generating high affinity ligands with prior knowledge of 3D target structures. Existing methods either use conditional generative model to learn the distribution of 3D ligands given target binding sites, or iteratively modify molecules to optimize a structure-based activity estimator. The former is highly constrained by data quantity and quality, which leaves optimization-based approaches more promising in practical scenario. However, existing optimization-based approaches choose to edit molecules in 2D space, and use molecular docking to estimate the activity using docking predicted 3D target-ligand complexes. The misalignment between the action space and the objective hinders the performance of these models, especially for those employ deep learning for acceleration. In this work, we propose MolEdit3D to combine 3D molecular generation with optimization frameworks. We develop a novel 3D graph editing model to generate molecules using fragments, and pre-train this model on abundant 3D ligands for learning target-independent properties. Then we employ a target-guided self-learning strategy to improve target-related properties using self-sampled molecules. MolEdit3D achieves state-of-the-art performance on majority of the evaluation metrics, and demonstrate strong capability of capturing both target-dependent and -independent properties.
MARS: Markov Molecular Sampling for Multi-objective Drug Discovery
Mar 18, 2021



Abstract:Searching for novel molecules with desired chemical properties is crucial in drug discovery. Existing work focuses on developing neural models to generate either molecular sequences or chemical graphs. However, it remains a big challenge to find novel and diverse compounds satisfying several properties. In this paper, we propose MARS, a method for multi-objective drug molecule discovery. MARS is based on the idea of generating the chemical candidates by iteratively editing fragments of molecular graphs. To search for high-quality candidates, it employs Markov chain Monte Carlo sampling (MCMC) on molecules with an annealing scheme and an adaptive proposal. To further improve sample efficiency, MARS uses a graph neural network (GNN) to represent and select candidate edits, where the GNN is trained on-the-fly with samples from MCMC. Experiments show that MARS achieves state-of-the-art performance in various multi-objective settings where molecular bio-activity, drug-likeness, and synthesizability are considered. Remarkably, in the most challenging setting where all four objectives are simultaneously optimized, our approach outperforms previous methods significantly in comprehensive evaluations. The code is available at https://github.com/yutxie/mars.
A New Local Transformation Module for Few-shot Segmentation
Nov 12, 2019



Abstract:Few-shot segmentation segments object regions of new classes with a few of manual annotations. Its key step is to establish the transformation module between support images (annotated images) and query images (unlabeled images), so that the segmentation cues of support images can guide the segmentation of query images. The existing methods form transformation model based on global cues, which however ignores the local cues that are verified in this paper to be very important for the transformation. This paper proposes a new transformation module based on local cues, where the relationship of the local features is used for transformation. To enhance the generalization performance of the network, the relationship matrix is calculated in a high-dimensional metric embedding space based on cosine distance. In addition, to handle the challenging mapping problem from the low-level local relationships to high-level semantic cues, we propose to apply generalized inverse matrix of the annotation matrix of support images to transform the relationship matrix linearly, which is non-parametric and class-agnostic. The result by the matrix transformation can be regarded as an attention map with high-level semantic cues, based on which a transformation module can be built simply.The proposed transformation module is a general module that can be used to replace the transformation module in the existing few-shot segmentation frameworks. We verify the effectiveness of the proposed method on Pascal VOC 2012 dataset. The value of mIoU achieves at 57.0% in 1-shot and 60.6% in 5-shot, which outperforms the state-of-the-art method by 1.6% and 3.5%, respectively.
A New Few-shot Segmentation Network Based on Class Representation
Sep 19, 2019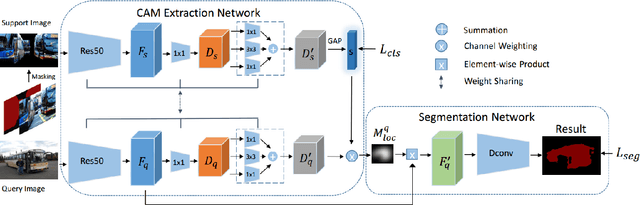
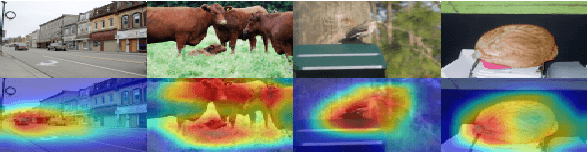

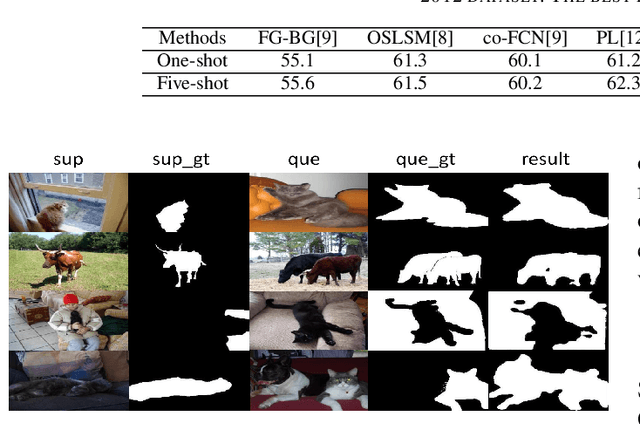
Abstract:This paper studies few-shot segmentation, which is a task of predicting foreground mask of unseen classes by a few of annotations only, aided by a set of rich annotations already existed. The existing methods mainly focus the task on "\textit{how to transfer segmentation cues from support images (labeled images) to query images (unlabeled images)}", and try to learn efficient and general transfer module that can be easily extended to unseen classes. However, it is proved to be a challenging task to learn the transfer module that is general to various classes. This paper solves few-shot segmentation in a new perspective of "\textit{how to represent unseen classes by existing classes}", and formulates few-shot segmentation as the representation process that represents unseen classes (in terms of forming the foreground prior) by existing classes precisely. Based on such idea, we propose a new class representation based few-shot segmentation framework, which firstly generates class activation map of unseen class based on the knowledge of existing classes, and then uses the map as foreground probability map to extract the foregrounds from query image. A new two-branch based few-shot segmentation network is proposed. Moreover, a new CAM generation module that extracts the CAM of unseen classes rather than the classical training classes is raised. We validate the effectiveness of our method on Pascal VOC 2012 dataset, the value FB-IoU of one-shot and five-shot arrives at 69.2\% and 70.1\% respectively, which outperforms the state-of-the-art method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge