Richard Chen
ContraGS: Codebook-Condensed and Trainable Gaussian Splatting for Fast, Memory-Efficient Reconstruction
Sep 03, 2025Abstract:3D Gaussian Splatting (3DGS) is a state-of-art technique to model real-world scenes with high quality and real-time rendering. Typically, a higher quality representation can be achieved by using a large number of 3D Gaussians. However, using large 3D Gaussian counts significantly increases the GPU device memory for storing model parameters. A large model thus requires powerful GPUs with high memory capacities for training and has slower training/rendering latencies due to the inefficiencies of memory access and data movement. In this work, we introduce ContraGS, a method to enable training directly on compressed 3DGS representations without reducing the Gaussian Counts, and thus with a little loss in model quality. ContraGS leverages codebooks to compactly store a set of Gaussian parameter vectors throughout the training process, thereby significantly reducing memory consumption. While codebooks have been demonstrated to be highly effective at compressing fully trained 3DGS models, directly training using codebook representations is an unsolved challenge. ContraGS solves the problem of learning non-differentiable parameters in codebook-compressed representations by posing parameter estimation as a Bayesian inference problem. To this end, ContraGS provides a framework that effectively uses MCMC sampling to sample over a posterior distribution of these compressed representations. With ContraGS, we demonstrate that ContraGS significantly reduces the peak memory during training (on average 3.49X) and accelerated training and rendering (1.36X and 1.88X on average, respectively), while retraining close to state-of-art quality.
Modeling Dense Multimodal Interactions Between Biological Pathways and Histology for Survival Prediction
Apr 13, 2023



Abstract:Integrating whole-slide images (WSIs) and bulk transcriptomics for predicting patient survival can improve our understanding of patient prognosis. However, this multimodal task is particularly challenging due to the different nature of these data: WSIs represent a very high-dimensional spatial description of a tumor, while bulk transcriptomics represent a global description of gene expression levels within that tumor. In this context, our work aims to address two key challenges: (1) how can we tokenize transcriptomics in a semantically meaningful and interpretable way?, and (2) how can we capture dense multimodal interactions between these two modalities? Specifically, we propose to learn biological pathway tokens from transcriptomics that can encode specific cellular functions. Together with histology patch tokens that encode the different morphological patterns in the WSI, we argue that they form appropriate reasoning units for downstream interpretability analyses. We propose fusing both modalities using a memory-efficient multimodal Transformer that can model interactions between pathway and histology patch tokens. Our proposed model, SURVPATH, achieves state-of-the-art performance when evaluated against both unimodal and multimodal baselines on five datasets from The Cancer Genome Atlas. Our interpretability framework identifies key multimodal prognostic factors, and, as such, can provide valuable insights into the interaction between genotype and phenotype, enabling a deeper understanding of the underlying biological mechanisms at play. We make our code public at: https://github.com/ajv012/SurvPath.
Quantum Deep Hedging
Mar 29, 2023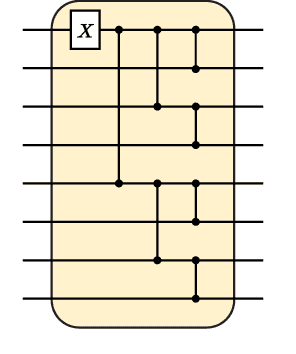
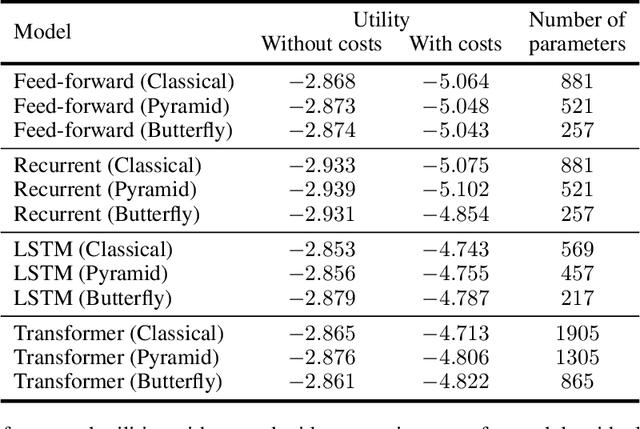
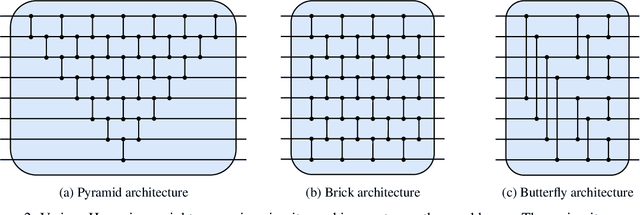
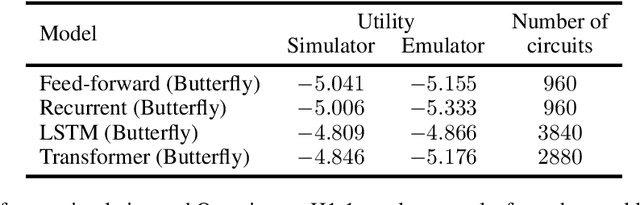
Abstract:Quantum machine learning has the potential for a transformative impact across industry sectors and in particular in finance. In our work we look at the problem of hedging where deep reinforcement learning offers a powerful framework for real markets. We develop quantum reinforcement learning methods based on policy-search and distributional actor-critic algorithms that use quantum neural network architectures with orthogonal and compound layers for the policy and value functions. We prove that the quantum neural networks we use are trainable, and we perform extensive simulations that show that quantum models can reduce the number of trainable parameters while achieving comparable performance and that the distributional approach obtains better performance than other standard approaches, both classical and quantum. We successfully implement the proposed models on a trapped-ion quantum processor, utilizing circuits with up to $16$ qubits, and observe performance that agrees well with noiseless simulation. Our quantum techniques are general and can be applied to other reinforcement learning problems beyond hedging.
Quantifying & Modeling Feature Interactions: An Information Decomposition Framework
Feb 23, 2023Abstract:The recent explosion of interest in multimodal applications has resulted in a wide selection of datasets and methods for representing and integrating information from different signals. Despite these empirical advances, there remain fundamental research questions: how can we quantify the nature of interactions that exist among input features? Subsequently, how can we capture these interactions using suitable data-driven methods? To answer this question, we propose an information-theoretic approach to quantify the degree of redundancy, uniqueness, and synergy across input features, which we term the PID statistics of a multimodal distribution. Using 2 newly proposed estimators that scale to high-dimensional distributions, we demonstrate their usefulness in quantifying the interactions within multimodal datasets, the nature of interactions captured by multimodal models, and principled approaches for model selection. We conduct extensive experiments on both synthetic datasets where the PID statistics are known and on large-scale multimodal benchmarks where PID estimation was previously impossible. Finally, to demonstrate the real-world applicability of our approach, we present three case studies in pathology, mood prediction, and robotic perception where our framework accurately recommends strong multimodal models for each application.
COVID-19 in differential diagnosis of online symptom assessments
Aug 07, 2020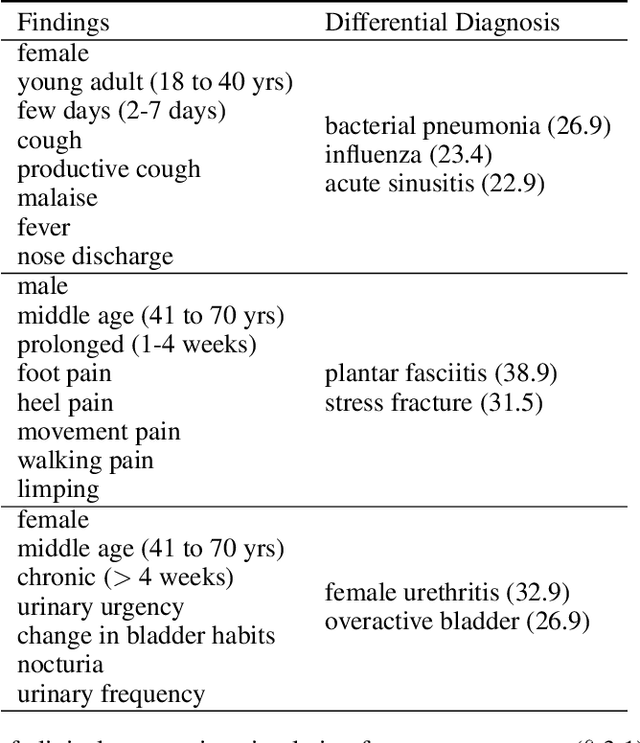
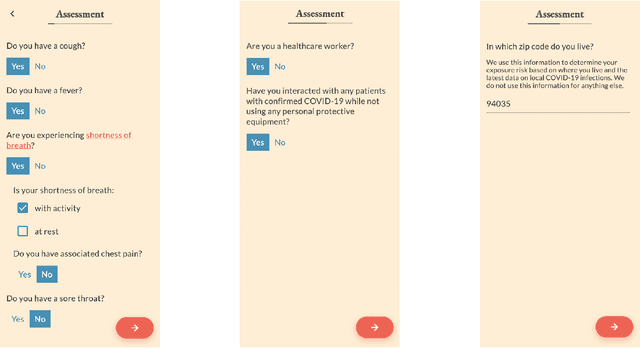

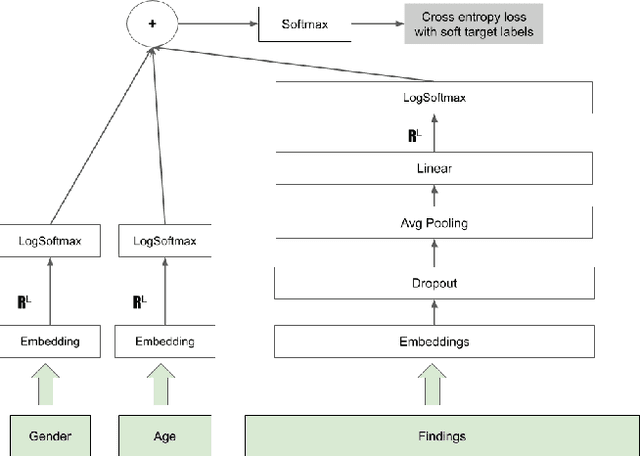
Abstract:The COVID-19 pandemic has magnified an already existing trend of people looking for healthcare solutions online. One class of solutions are symptom checkers, which have become very popular in the context of COVID-19. Traditional symptom checkers, however, are based on manually curated expert systems that are inflexible and hard to modify, especially in a quickly changing situation like the one we are facing today. That is why all COVID-19 existing solutions are manual symptom checkers that can only estimate the probability of this disease and cannot contemplate alternative hypothesis or come up with a differential diagnosis. While machine learning offers an alternative, the lack of reliable data does not make it easy to apply to COVID-19 either. In this paper we present an approach that combines the strengths of traditional AI expert systems and novel deep learning models. In doing so we can leverage prior knowledge as well as any amount of existing data to quickly derive models that best adapt to the current state of the world and latest scientific knowledge. We use the approach to train a COVID-19 aware differential diagnosis model that can be used for medical decision support both for doctors or patients. We show that our approach is able to accurately model new incoming data about COVID-19 while still preserving accuracy on conditions that had been modeled in the past. While our approach shows evident and clear advantages for an extreme situation like the one we are currently facing, we also show that its flexibility generalizes beyond this concrete, but very important, example.
A Domain-agnostic, Noise-resistant, Hardware-efficient Evolutionary Variational Quantum Eigensolver
Nov 18, 2019



Abstract:Variational quantum algorithms have shown promise in numerous fields due to their versatility in solving problems of scientific and commercial interest. However, leading algorithms for Hamiltonian simulation, such as the Variational Quantum Eigensolver (VQE), use fixed preconstructed ansatzes, limiting their general applicability and accuracy. Thus, variational forms---the quantum circuits that implement ansatzes ---are either crafted heuristically or by encoding domain-specific knowledge. In this paper, we present an Evolutionary Variational Quantum Eigensolver (EVQE), a novel variational algorithm that uses evolutionary programming techniques to minimize the expectation value of a given Hamiltonian by dynamically generating and optimizing an ansatz. The algorithm is equally applicable to optimization problems in all domains, obtaining accurate energy evaluations with hardware-efficient ansatzes. In molecular simulations, the variational forms generated by EVQE are up to $18.6\times$ shallower and use up to $12\times$ fewer CX gates than those obtained by VQE with a unitary coupled cluster ansatz. EVQE demonstrates significant noise-resistance properties, obtaining results in noisy simulation with at least $3.6\times$ less error than VQE using any tested ansatz configuration. We successfully evaluated EVQE on a real 5-qubit IBMQ quantum computer. The experimental results, which we obtained both via simulation and on real quantum hardware, demonstrate the effectiveness of EVQE for general-purpose optimization on the quantum computers of the present and near future.
Learning Montezuma's Revenge from a Single Demonstration
Dec 08, 2018



Abstract:We propose a new method for learning from a single demonstration to solve hard exploration tasks like the Atari game Montezuma's Revenge. Instead of imitating human demonstrations, as proposed in other recent works, our approach is to maximize rewards directly. Our agent is trained using off-the-shelf reinforcement learning, but starts every episode by resetting to a state from a demonstration. By starting from such demonstration states, the agent requires much less exploration to learn a game compared to when it starts from the beginning of the game at every episode. We analyze reinforcement learning for tasks with sparse rewards in a simple toy environment, where we show that the run-time of standard RL methods scales exponentially in the number of states between rewards. Our method reduces this to quadratic scaling, opening up many tasks that were previously infeasible. We then apply our method to Montezuma's Revenge, for which we present a trained agent achieving a high-score of 74,500, better than any previously published result.
Deep Adversarial Training for Multi-Organ Nuclei Segmentation in Histopathology Images
Oct 19, 2018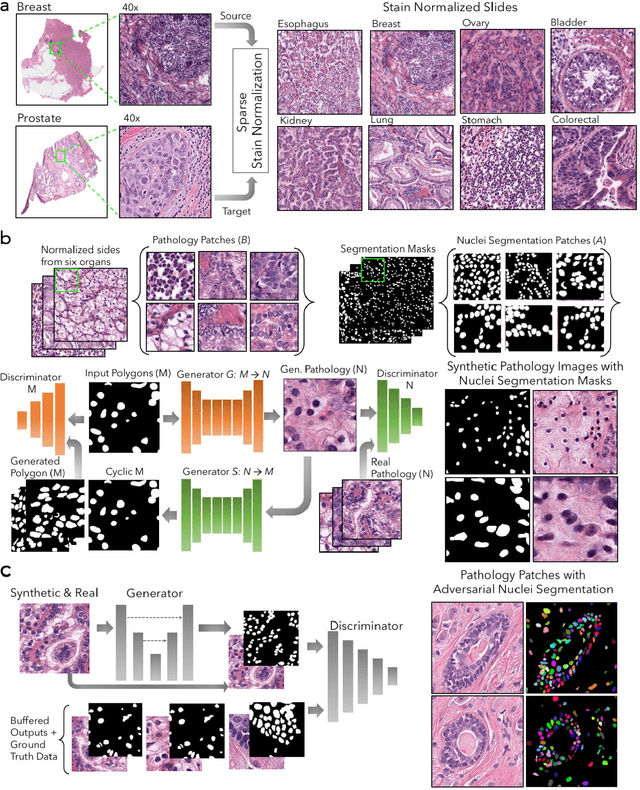
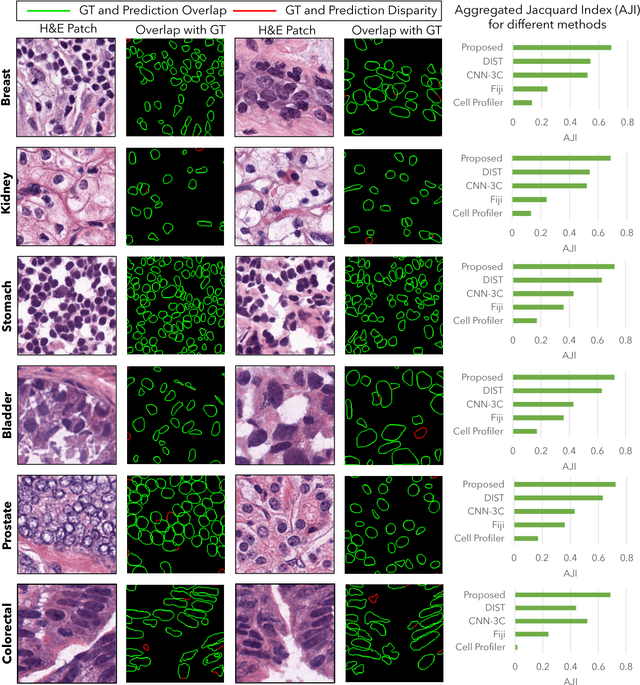
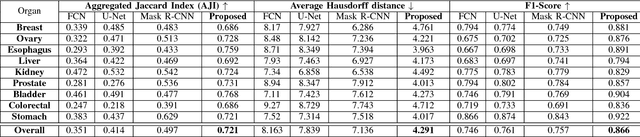
Abstract:Nuclei segmentation is a fundamental task that is critical for various computational pathology applications including nuclei morphology analysis, cell type classification, and cancer grading. Conventional vision-based methods for nuclei segmentation struggle in challenging cases and deep learning approaches have proven to be more robust and generalizable. However, CNNs require large amounts of labeled histopathology data. Moreover, conventional CNN-based approaches lack structured prediction capabilities which are required to distinguish overlapping and clumped nuclei. Here, we present an approach to nuclei segmentation that overcomes these challenges by utilizing a conditional generative adversarial network (cGAN) trained with synthetic and real data. We generate a large dataset of H&E training images with perfect nuclei segmentation labels using an unpaired GAN framework. This synthetic data along with real histopathology data from six different organs are used to train a conditional GAN with spectral normalization and gradient penalty for nuclei segmentation. This adversarial regression framework enforces higher order consistency when compared to conventional CNN models. We demonstrate that this nuclei segmentation approach generalizes across different organs, sites, patients and disease states, and outperforms conventional approaches, especially in isolating individual and overlapping nuclei.
Deep Learning with Cinematic Rendering: Fine-Tuning Deep Neural Networks Using Photorealistic Medical Images
Sep 29, 2018



Abstract:Deep learning has emerged as a powerful artificial intelligence tool to interpret medical images for a growing variety of applications. However, the paucity of medical imaging data with high-quality annotations that is necessary for training such methods ultimately limits their performance. Medical data is challenging to acquire due to privacy issues, shortage of experts available for annotation, limited representation of rare conditions and cost. This problem has previously been addressed by using synthetically generated data. However, networks trained on synthetic data often fail to generalize to real data. Cinematic rendering simulates the propagation and interaction of light passing through tissue models reconstructed from CT data, enabling the generation of photorealistic images. In this paper, we present one of the first applications of cinematic rendering in deep learning, in which we propose to fine-tune synthetic data-driven networks using cinematically rendered CT data for the task of monocular depth estimation in endoscopy. Our experiments demonstrate that: (a) Convolutional Neural Networks (CNNs) trained on synthetic data and fine-tuned on photorealistic cinematically rendered data adapt better to real medical images and demonstrate more robust performance when compared to networks with no fine-tuning, (b) these fine-tuned networks require less training data to converge to an optimal solution, and (c) fine-tuning with data from a variety of photorealistic rendering conditions of the same scene prevents the network from learning patient-specific information and aids in generalizability of the model. Our empirical evaluation demonstrates that networks fine-tuned with cinematically rendered data predict depth with 56.87% less error for rendered endoscopy images and 27.49% less error for real porcine colon endoscopy images.
Rethinking Monocular Depth Estimation with Adversarial Training
Sep 24, 2018



Abstract:Monocular depth estimation is an extensively studied computer vision problem with a vast variety of applications. Deep learning-based methods have demonstrated promise for both supervised and unsupervised depth estimation from monocular images. Most existing approaches treat depth estimation as a regression problem with a local pixel-wise loss function. In this work, we innovate beyond existing approaches by using adversarial training to learn a context-aware, non-local loss function. Such an approach penalizes the joint configuration of predicted depth values at the patch-level instead of the pixel-level, which allows networks to incorporate more global information. In this framework, the generator learns a mapping between RGB images and its corresponding depth map, while the discriminator learns to distinguish depth map and RGB pairs from ground truth. This conditional GAN depth estimation framework is stabilized using spectral normalization to prevent mode collapse when learning from diverse datasets. We test this approach using a diverse set of generators that include U-Net and joint CNN-CRF. We benchmark this approach on the NYUv2, Make3D and KITTI datasets, and observe that adversarial training reduces relative error by several fold, achieving state-of-the-art performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge