Xavier Amatriain
Large Language Models: A Survey
Feb 20, 2024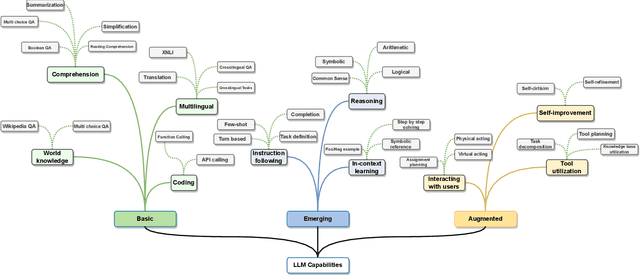
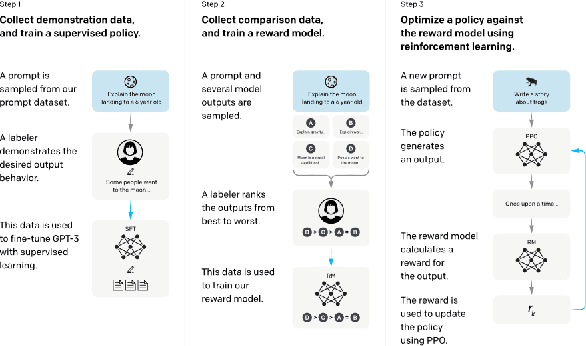
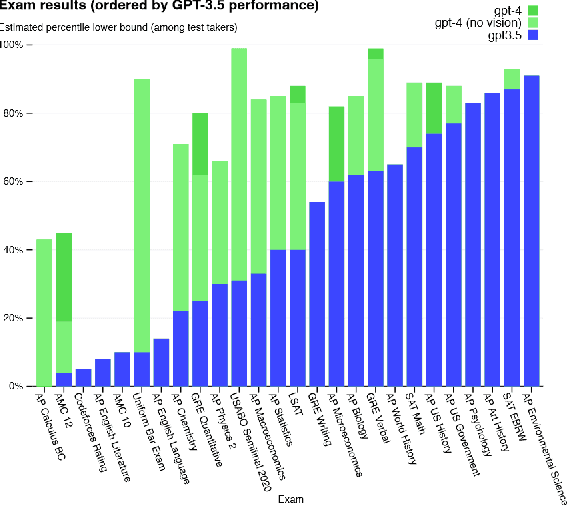
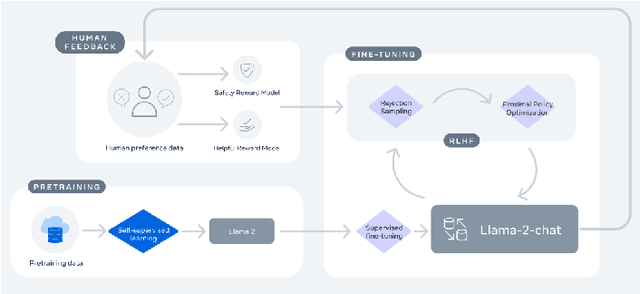
Abstract:Large Language Models (LLMs) have drawn a lot of attention due to their strong performance on a wide range of natural language tasks, since the release of ChatGPT in November 2022. LLMs' ability of general-purpose language understanding and generation is acquired by training billions of model's parameters on massive amounts of text data, as predicted by scaling laws \cite{kaplan2020scaling,hoffmann2022training}. The research area of LLMs, while very recent, is evolving rapidly in many different ways. In this paper, we review some of the most prominent LLMs, including three popular LLM families (GPT, LLaMA, PaLM), and discuss their characteristics, contributions and limitations. We also give an overview of techniques developed to build, and augment LLMs. We then survey popular datasets prepared for LLM training, fine-tuning, and evaluation, review widely used LLM evaluation metrics, and compare the performance of several popular LLMs on a set of representative benchmarks. Finally, we conclude the paper by discussing open challenges and future research directions.
Prompt Design and Engineering: Introduction and Advanced Methods
Jan 30, 2024Abstract:Prompt design and engineering has become an important discipline in just the past few months. In this paper, we provide an introduction to the main concepts and design approaches. We also provide more advanced techniques all the way to those needed to design LLM-based agents. We finish by providing a list of existing tools for prompt engineering.
Injecting knowledge into language generation: a case study in auto-charting after-visit care instructions from medical dialogue
Jun 06, 2023



Abstract:Factual correctness is often the limiting factor in practical applications of natural language generation in high-stakes domains such as healthcare. An essential requirement for maintaining factuality is the ability to deal with rare tokens. This paper focuses on rare tokens that appear in both the source and the reference sequences, and which, when missed during generation, decrease the factual correctness of the output text. For high-stake domains that are also knowledge-rich, we show how to use knowledge to (a) identify which rare tokens that appear in both source and reference are important and (b) uplift their conditional probability. We introduce the ``utilization rate'' that encodes knowledge and serves as a regularizer by maximizing the marginal probability of selected tokens. We present a study in a knowledge-rich domain of healthcare, where we tackle the problem of generating after-visit care instructions based on patient-doctor dialogues. We verify that, in our dataset, specific medical concepts with high utilization rates are underestimated by conventionally trained sequence-to-sequence models. We observe that correcting this with our approach to knowledge injection reduces the uncertainty of the model as well as improves factuality and coherence without negatively impacting fluency.
Transformer models: an introduction and catalog
Feb 16, 2023Abstract:In the past few years we have seen the meteoric appearance of dozens of models of the Transformer family, all of which have funny, but not self-explanatory, names. The goal of this paper is to offer a somewhat comprehensive but simple catalog and classification of the most popular Transformer models. The paper also includes an introduction to the most important aspects and innovation in Transformer models.
Learning functional sections in medical conversations: iterative pseudo-labeling and human-in-the-loop approach
Oct 07, 2022



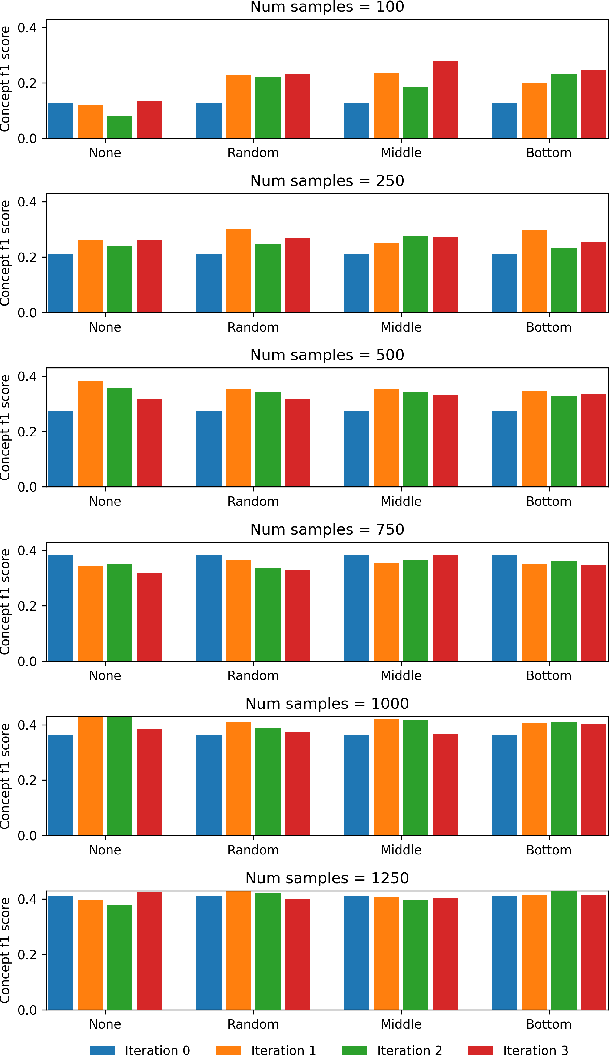
Abstract:Medical conversations between patients and medical professionals have implicit functional sections, such as "history taking", "summarization", "education", and "care plan." In this work, we are interested in learning to automatically extract these sections. A direct approach would require collecting large amounts of expert annotations for this task, which is inherently costly due to the contextual inter-and-intra variability between these sections. This paper presents an approach that tackles the problem of learning to classify medical dialogue into functional sections without requiring a large number of annotations. Our approach combines pseudo-labeling and human-in-the-loop. First, we bootstrap using weak supervision with pseudo-labeling to generate dialogue turn-level pseudo-labels and train a transformer-based model, which is then applied to individual sentences to create noisy sentence-level labels. Second, we iteratively refine sentence-level labels using a cluster-based human-in-the-loop approach. Each iteration requires only a few dozen annotator decisions. We evaluate the results on an expert-annotated dataset of 100 dialogues and find that while our models start with 69.5% accuracy, we can iteratively improve it to 82.5%. The code used to perform all experiments described in this paper can be found here: https://github.com/curai/curai-research/tree/main/functional-sections.
OSLAT: Open Set Label Attention Transformer for Medical Entity Span Extraction
Jul 12, 2022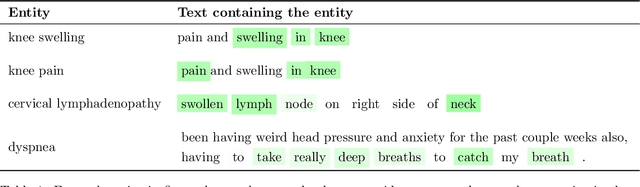
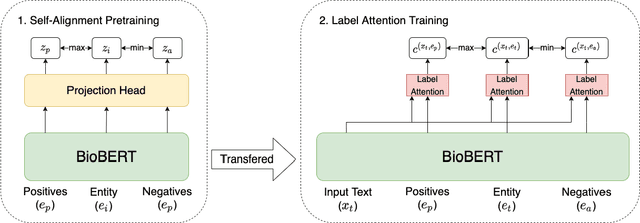
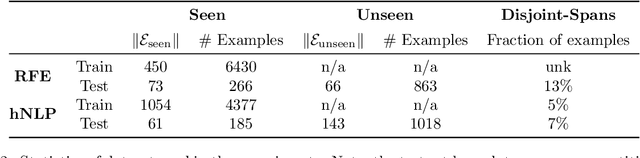
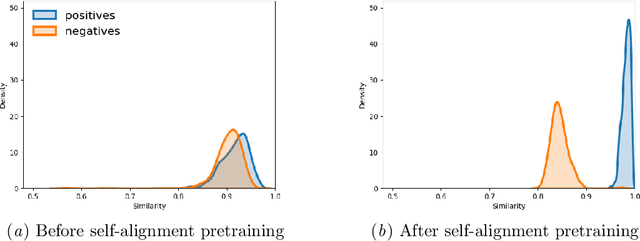
Abstract:Identifying spans in medical texts that correspond to medical entities is one of the core steps for many healthcare NLP tasks such as ICD coding, medical finding extraction, medical note contextualization, to name a few. Existing entity extraction methods rely on a fixed and limited vocabulary of medical entities and have difficulty with extracting entities represented by disjoint spans. In this paper, we present a new transformer-based architecture called OSLAT, Open Set Label Attention Transformer, that addresses many of the limitations of the previous methods. Our approach uses the label-attention mechanism to implicitly learn spans associated with entities of interest. These entities can be provided as free text, including entities not seen during OSLAT's training, and the model can extract spans even when they are disjoint. To test the generalizability of our method, we train two separate models on two different datasets, which have very low entity overlap: (1) a public discharge notes dataset from hNLP, and (2) a much more challenging proprietary patient text dataset "Reasons for Encounter" (RFE). We find that OSLAT models trained on either dataset outperform rule-based and fuzzy string matching baselines when applied to the RFE dataset as well as to the portion of hNLP dataset where entities are represented by disjoint spans. Our code can be found at https://github.com/curai/curai-research/tree/main/OSLAT.
Adding more data does not always help: A study in medical conversation summarization with PEGASUS
Nov 28, 2021

Abstract:Medical conversation summarization is integral in capturing information gathered during interactions between patients and physicians. Summarized conversations are used to facilitate patient hand-offs between physicians, and as part of providing care in the future. Summaries, however, can be time-consuming to produce and require domain expertise. Modern pre-trained NLP models such as PEGASUS have emerged as capable alternatives to human summarization, reaching state-of-the-art performance on many summarization benchmarks. However, many downstream tasks still require at least moderately sized datasets to achieve satisfactory performance. In this work we (1) explore the effect of dataset size on transfer learning medical conversation summarization using PEGASUS and (2) evaluate various iterative labeling strategies in the low-data regime, following their success in the classification setting. We find that model performance saturates with increase in dataset size and that the various active-learning strategies evaluated all show equivalent performance consistent with simple dataset size increase. We also find that naive iterative pseudo-labeling is on-par or slightly worse than no pseudo-labeling. Our work sheds light on the successes and challenges of translating low-data regime techniques in classification to medical conversation summarization and helps guides future work in this space. Relevant code available at \url{https://github.com/curai/curai-research/tree/main/medical-summarization-ML4H-2021}.
MEDCOD: A Medically-Accurate, Emotive, Diverse, and Controllable Dialog System
Nov 17, 2021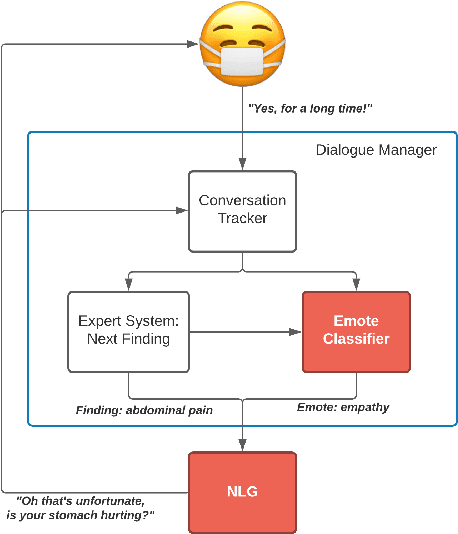


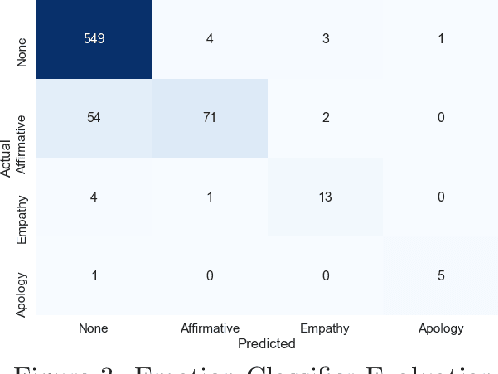
Abstract:We present MEDCOD, a Medically-Accurate, Emotive, Diverse, and Controllable Dialog system with a unique approach to the natural language generator module. MEDCOD has been developed and evaluated specifically for the history taking task. It integrates the advantage of a traditional modular approach to incorporate (medical) domain knowledge with modern deep learning techniques to generate flexible, human-like natural language expressions. Two key aspects of MEDCOD's natural language output are described in detail. First, the generated sentences are emotive and empathetic, similar to how a doctor would communicate to the patient. Second, the generated sentence structures and phrasings are varied and diverse while maintaining medical consistency with the desired medical concept (provided by the dialogue manager module of MEDCOD). Experimental results demonstrate the effectiveness of our approach in creating a human-like medical dialogue system. Relevant code is available at https://github.com/curai/curai-research/tree/main/MEDCOD
Medical symptom recognition from patient text: An active learning approach for long-tailed multilabel distributions
Nov 12, 2020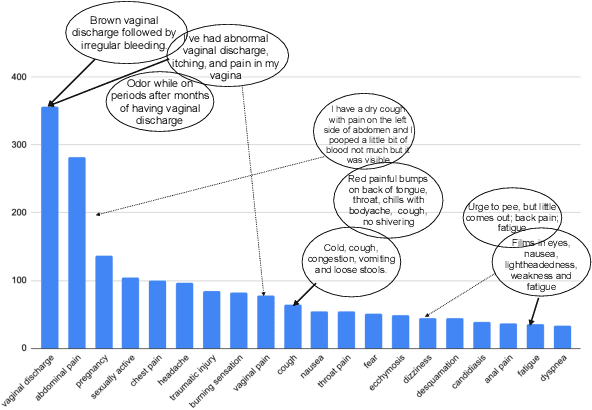


Abstract:We study the problem of medical symptoms recognition from patient text, for the purposes of gathering pertinent information from the patient (known as history-taking). We introduce an active learning method that leverages underlying structure of a continually refined, learned latent space to select the most informative examples to label. This enables the selection of the most informative examples that progressively increases the coverage on the universe of symptoms via the learned model, despite the long tail in data distribution.
Dr. Summarize: Global Summarization of Medical Dialogue by Exploiting Local Structures
Sep 18, 2020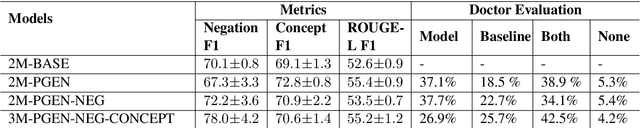
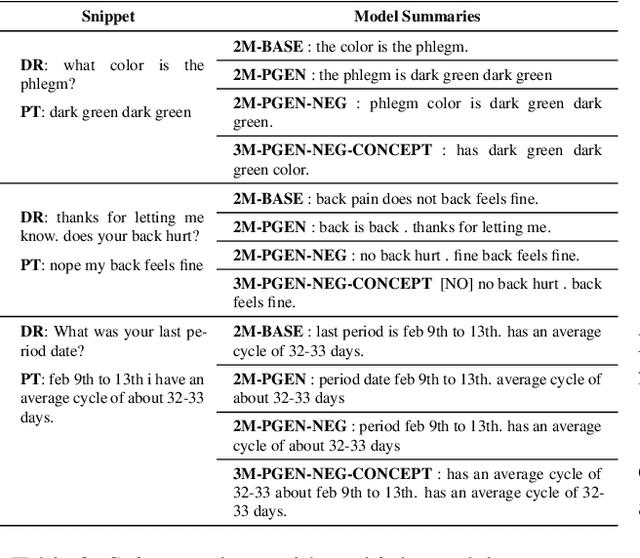
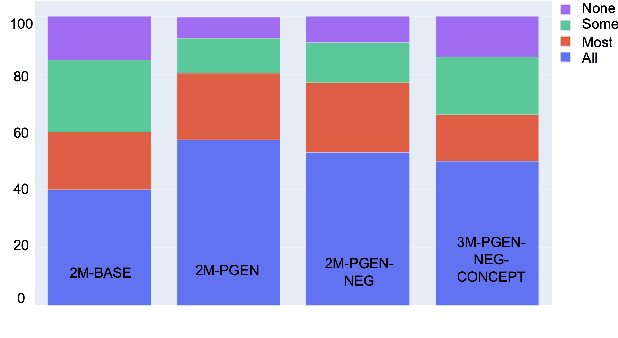
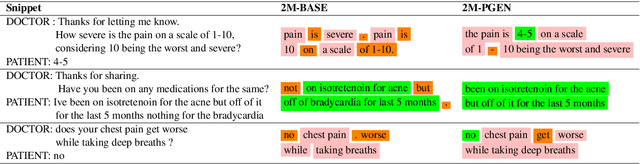
Abstract:Understanding a medical conversation between a patient and a physician poses a unique natural language understanding challenge since it combines elements of standard open ended conversation with very domain specific elements that require expertise and medical knowledge. Summarization of medical conversations is a particularly important aspect of medical conversation understanding since it addresses a very real need in medical practice: capturing the most important aspects of a medical encounter so that they can be used for medical decision making and subsequent follow ups. In this paper we present a novel approach to medical conversation summarization that leverages the unique and independent local structures created when gathering a patient's medical history. Our approach is a variation of the pointer generator network where we introduce a penalty on the generator distribution, and we explicitly model negations. The model also captures important properties of medical conversations such as medical knowledge coming from standardized medical ontologies better than when those concepts are introduced explicitly. Through evaluation by doctors, we show that our approach is preferred on twice the number of summaries to the baseline pointer generator model and captures most or all of the information in 80% of the conversations making it a realistic alternative to costly manual summarization by medical experts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge