Namit Katariya
Adding more data does not always help: A study in medical conversation summarization with PEGASUS
Nov 28, 2021

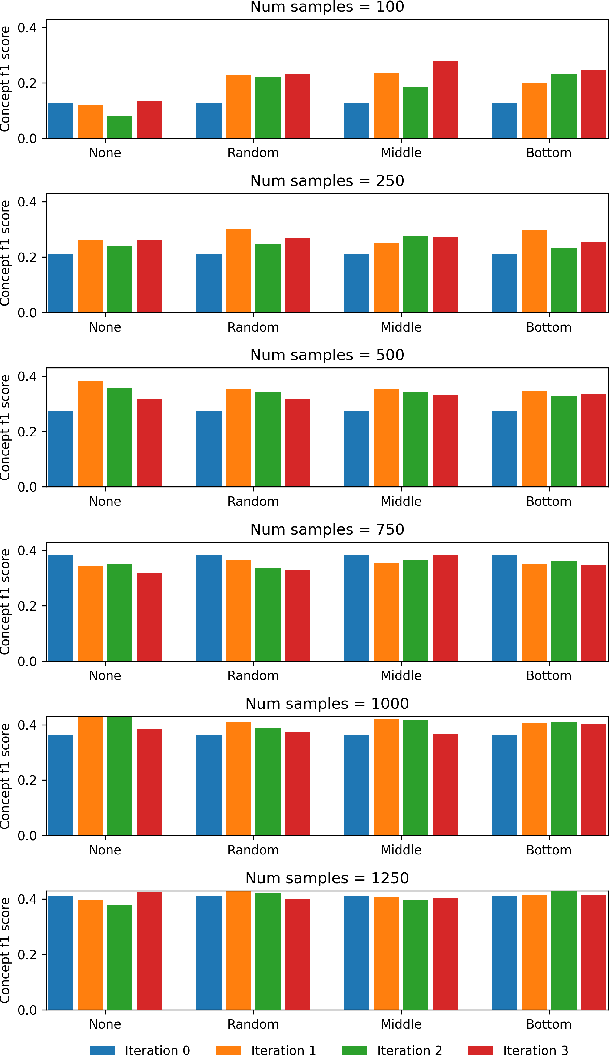
Abstract:Medical conversation summarization is integral in capturing information gathered during interactions between patients and physicians. Summarized conversations are used to facilitate patient hand-offs between physicians, and as part of providing care in the future. Summaries, however, can be time-consuming to produce and require domain expertise. Modern pre-trained NLP models such as PEGASUS have emerged as capable alternatives to human summarization, reaching state-of-the-art performance on many summarization benchmarks. However, many downstream tasks still require at least moderately sized datasets to achieve satisfactory performance. In this work we (1) explore the effect of dataset size on transfer learning medical conversation summarization using PEGASUS and (2) evaluate various iterative labeling strategies in the low-data regime, following their success in the classification setting. We find that model performance saturates with increase in dataset size and that the various active-learning strategies evaluated all show equivalent performance consistent with simple dataset size increase. We also find that naive iterative pseudo-labeling is on-par or slightly worse than no pseudo-labeling. Our work sheds light on the successes and challenges of translating low-data regime techniques in classification to medical conversation summarization and helps guides future work in this space. Relevant code available at \url{https://github.com/curai/curai-research/tree/main/medical-summarization-ML4H-2021}.
MEDCOD: A Medically-Accurate, Emotive, Diverse, and Controllable Dialog System
Nov 17, 2021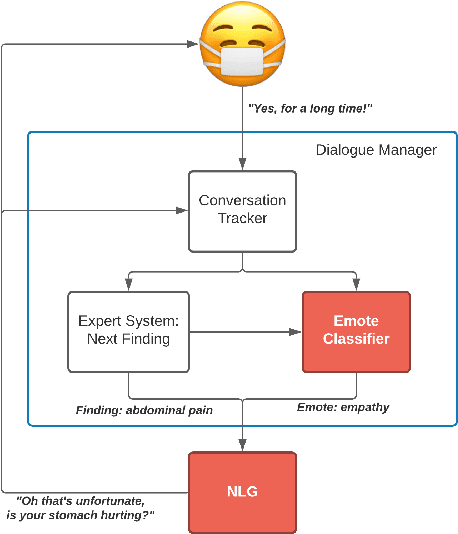


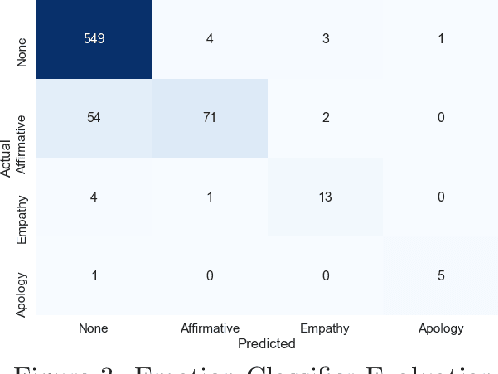
Abstract:We present MEDCOD, a Medically-Accurate, Emotive, Diverse, and Controllable Dialog system with a unique approach to the natural language generator module. MEDCOD has been developed and evaluated specifically for the history taking task. It integrates the advantage of a traditional modular approach to incorporate (medical) domain knowledge with modern deep learning techniques to generate flexible, human-like natural language expressions. Two key aspects of MEDCOD's natural language output are described in detail. First, the generated sentences are emotive and empathetic, similar to how a doctor would communicate to the patient. Second, the generated sentence structures and phrasings are varied and diverse while maintaining medical consistency with the desired medical concept (provided by the dialogue manager module of MEDCOD). Experimental results demonstrate the effectiveness of our approach in creating a human-like medical dialogue system. Relevant code is available at https://github.com/curai/curai-research/tree/main/MEDCOD
Dr. Summarize: Global Summarization of Medical Dialogue by Exploiting Local Structures
Sep 18, 2020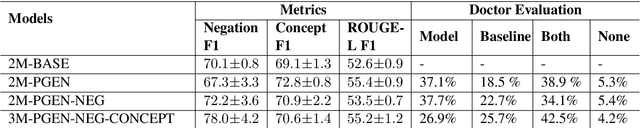
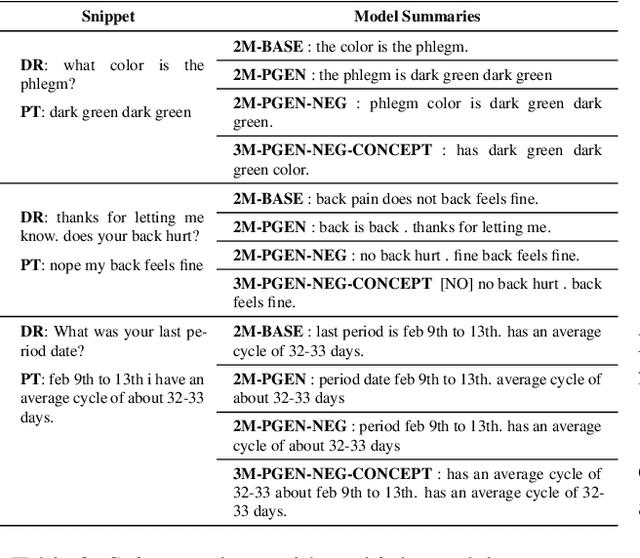
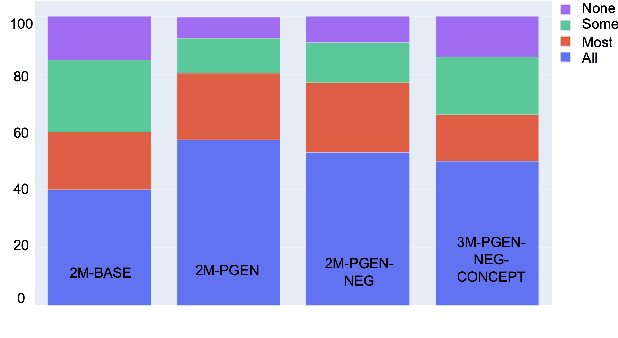
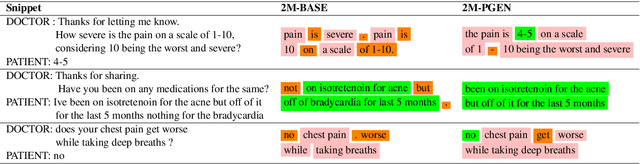
Abstract:Understanding a medical conversation between a patient and a physician poses a unique natural language understanding challenge since it combines elements of standard open ended conversation with very domain specific elements that require expertise and medical knowledge. Summarization of medical conversations is a particularly important aspect of medical conversation understanding since it addresses a very real need in medical practice: capturing the most important aspects of a medical encounter so that they can be used for medical decision making and subsequent follow ups. In this paper we present a novel approach to medical conversation summarization that leverages the unique and independent local structures created when gathering a patient's medical history. Our approach is a variation of the pointer generator network where we introduce a penalty on the generator distribution, and we explicitly model negations. The model also captures important properties of medical conversations such as medical knowledge coming from standardized medical ontologies better than when those concepts are introduced explicitly. Through evaluation by doctors, we show that our approach is preferred on twice the number of summaries to the baseline pointer generator model and captures most or all of the information in 80% of the conversations making it a realistic alternative to costly manual summarization by medical experts.
Effective Transfer Learning for Identifying Similar Questions: Matching User Questions to COVID-19 FAQs
Aug 04, 2020
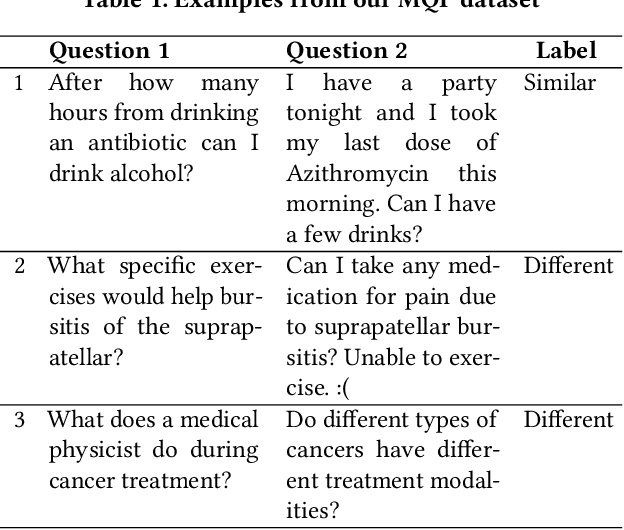
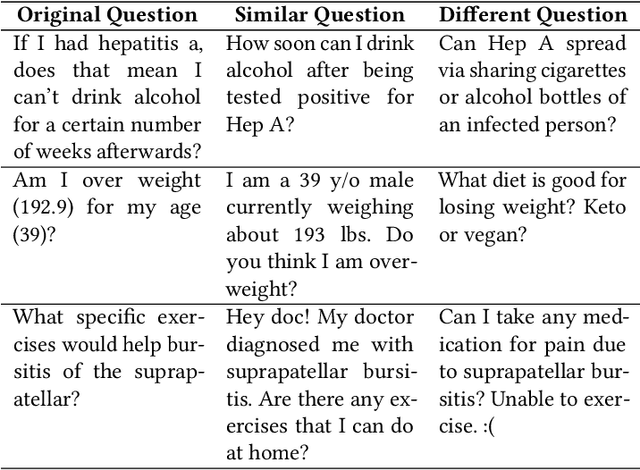
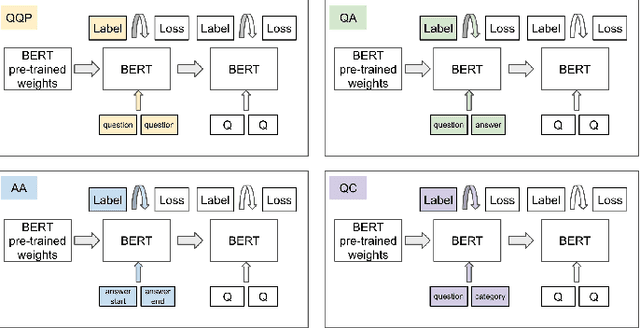
Abstract:People increasingly search online for answers to their medical questions but the rate at which medical questions are asked online significantly exceeds the capacity of qualified people to answer them. This leaves many questions unanswered or inadequately answered. Many of these questions are not unique, and reliable identification of similar questions would enable more efficient and effective question answering schema. COVID-19 has only exacerbated this problem. Almost every government agency and healthcare organization has tried to meet the informational need of users by building online FAQs, but there is no way for people to ask their question and know if it is answered on one of these pages. While many research efforts have focused on the problem of general question similarity, these approaches do not generalize well to domains that require expert knowledge to determine semantic similarity, such as the medical domain. In this paper, we show how a double fine-tuning approach of pretraining a neural network on medical question-answer pairs followed by fine-tuning on medical question-question pairs is a particularly useful intermediate task for the ultimate goal of determining medical question similarity. While other pretraining tasks yield an accuracy below 78.7% on this task, our model achieves an accuracy of 82.6% with the same number of training examples, an accuracy of 80.0% with a much smaller training set, and an accuracy of 84.5% when the full corpus of medical question-answer data is used. We also describe a currently live system that uses the trained model to match user questions to COVID-related FAQs.
Classification as Decoder: Trading Flexibility for Control in Medical Dialogue
Nov 16, 2019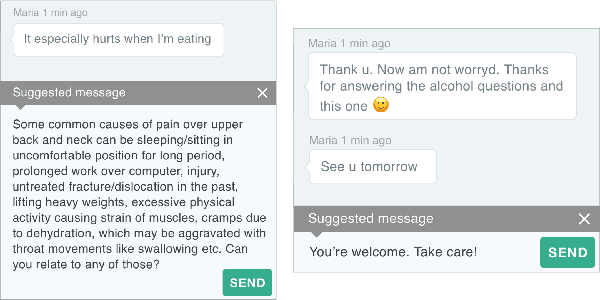
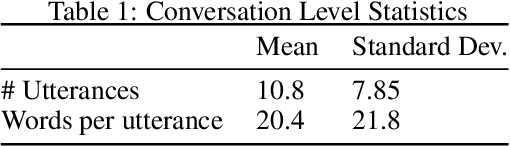
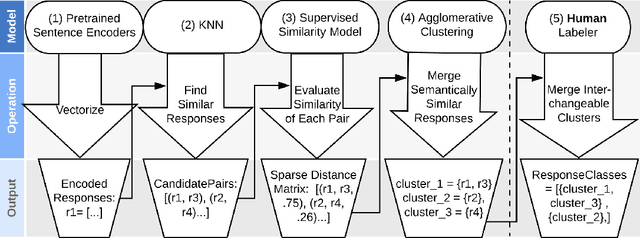
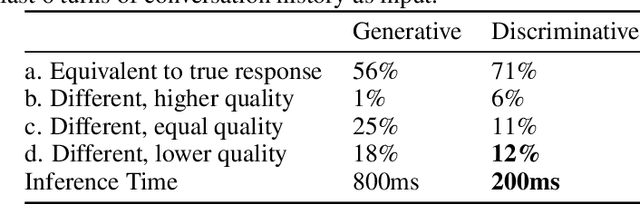
Abstract:Generative seq2seq dialogue systems are trained to predict the next word in dialogues that have already occurred. They can learn from large unlabeled conversation datasets, build a deeper understanding of conversational context, and generate a wide variety of responses. This flexibility comes at the cost of control, a concerning tradeoff in doctor/patient interactions. Inaccuracies, typos, or undesirable content in the training data will be reproduced by the model at inference time. We trade a small amount of labeling effort and some loss of response variety in exchange for quality control. More specifically, a pretrained language model encodes the conversational context, and we finetune a classification head to map an encoded conversational context to a response class, where each class is a noisily labeled group of interchangeable responses. Experts can update these exemplar responses over time as best practices change without retraining the classifier or invalidating old training data. Expert evaluation of 775 unseen doctor/patient conversations shows that only 12% of the discriminative model's responses are worse than the what the doctor ended up writing, compared to 18% for the generative model.
Domain-Relevant Embeddings for Medical Question Similarity
Nov 15, 2019
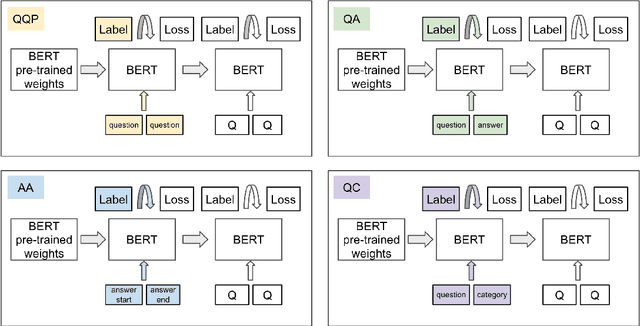

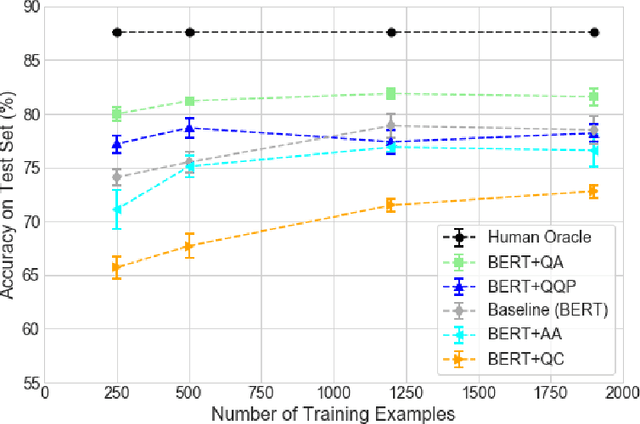
Abstract:The rate at which medical questions are asked online far exceeds the capacity of qualified people to answer them, and many of these questions are not unique. Identifying same-question pairs could enable questions to be answered more effectively. While many research efforts have focused on the problem of general question similarity for non-medical applications, these approaches do not generalize well to the medical domain, where medical expertise is often required to determine semantic similarity. In this paper, we show how a semi-supervised approach of pre-training a neural network on medical question-answer pairs is a particularly useful intermediate task for the ultimate goal of determining medical question similarity. While other pre-training tasks yield an accuracy below 78.7% on this task, our model achieves an accuracy of 82.6% with the same number of training examples, and an accuracy of 80.0% with a much smaller training set.
Classification As Decoder: Trading Flexibility For Control In Neural Dialogue
Oct 17, 2019



Abstract:Generative seq2seq dialogue systems are trained to predict the next word in dialogues that have already occurred. They can learn from large unlabeled conversation datasets, build a deep understanding of conversational context, and generate a wide variety of responses. This flexibility comes at the cost of control. Undesirable responses in the training data will be reproduced by the model at inference time, and longer generations often don't make sense. Instead of generating responses one word at a time, we train a classifier to choose from a predefined list of full responses. The classifier is trained on (conversation context, response class) pairs, where each response class is a noisily labeled group of interchangeable responses. At inference, we generate the exemplar response associated with the predicted response class. Experts can edit and improve these exemplar responses over time without retraining the classifier or invalidating old training data. Human evaluation of 775 unseen doctor/patient conversations shows that this tradeoff improves responses. Only 12% of our discriminative approach's responses are worse than the doctor's response in the same conversational context, compared to 18% for the generative model. A discriminative model trained without any manual labeling of response classes achieves equal performance to the generative model.
Open Set Medical Diagnosis
Oct 07, 2019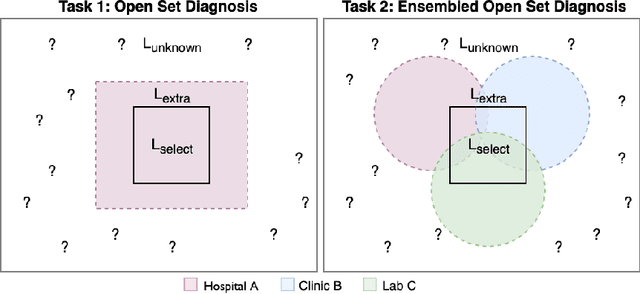
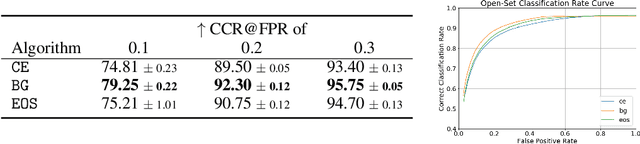
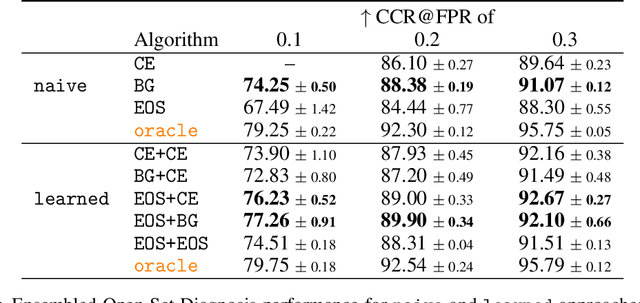

Abstract:Machine-learned diagnosis models have shown promise as medical aides but are trained under a closed-set assumption, i.e. that models will only encounter conditions on which they have been trained. However, it is practically infeasible to obtain sufficient training data for every human condition, and once deployed such models will invariably face previously unseen conditions. We frame machine-learned diagnosis as an open-set learning problem, and study how state-of-the-art approaches compare. Further, we extend our study to a setting where training data is distributed across several healthcare sites that do not allow data pooling, and experiment with different strategies of building open-set diagnostic ensembles. Across both settings, we observe consistent gains from explicitly modeling unseen conditions, but find the optimal training strategy to vary across settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge