Anitha Kannan
Rare Disease Differential Diagnosis with Large Language Models at Scale: From Abdominal Actinomycosis to Wilson's Disease
Feb 20, 2025



Abstract:Large language models (LLMs) have demonstrated impressive capabilities in disease diagnosis. However, their effectiveness in identifying rarer diseases, which are inherently more challenging to diagnose, remains an open question. Rare disease performance is critical with the increasing use of LLMs in healthcare settings. This is especially true if a primary care physician needs to make a rarer prognosis from only a patient conversation so that they can take the appropriate next step. To that end, several clinical decision support systems are designed to support providers in rare disease identification. Yet their utility is limited due to their lack of knowledge of common disorders and difficulty of use. In this paper, we propose RareScale to combine the knowledge LLMs with expert systems. We use jointly use an expert system and LLM to simulate rare disease chats. This data is used to train a rare disease candidate predictor model. Candidates from this smaller model are then used as additional inputs to black-box LLM to make the final differential diagnosis. Thus, RareScale allows for a balance between rare and common diagnoses. We present results on over 575 rare diseases, beginning with Abdominal Actinomycosis and ending with Wilson's Disease. Our approach significantly improves the baseline performance of black-box LLMs by over 17% in Top-5 accuracy. We also find that our candidate generation performance is high (e.g. 88.8% on gpt-4o generated chats).
Extrinsically-Focused Evaluation of Omissions in Medical Summarization
Nov 14, 2023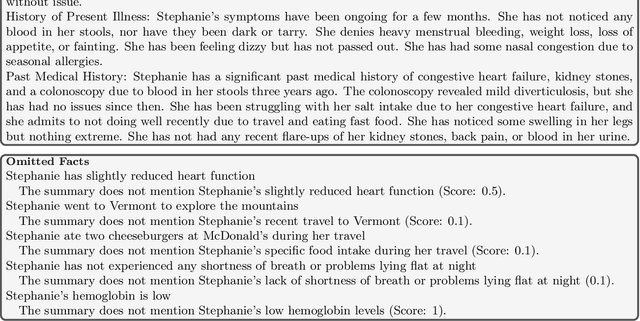

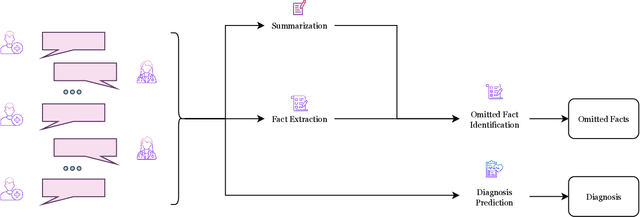
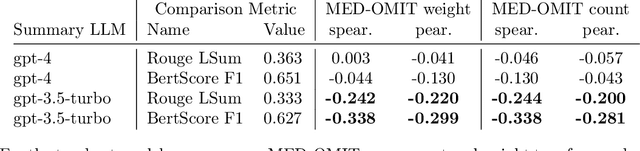
Abstract:The goal of automated summarization techniques (Paice, 1990; Kupiec et al, 1995) is to condense text by focusing on the most critical information. Generative large language models (LLMs) have shown to be robust summarizers, yet traditional metrics struggle to capture resulting performance (Goyal et al, 2022) in more powerful LLMs. In safety-critical domains such as medicine, more rigorous evaluation is required, especially given the potential for LLMs to omit important information in the resulting summary. We propose MED-OMIT, a new omission benchmark for medical summarization. Given a doctor-patient conversation and a generated summary, MED-OMIT categorizes the chat into a set of facts and identifies which are omitted from the summary. We further propose to determine fact importance by simulating the impact of each fact on a downstream clinical task: differential diagnosis (DDx) generation. MED-OMIT leverages LLM prompt-based approaches which categorize the importance of facts and cluster them as supporting or negating evidence to the diagnosis. We evaluate MED-OMIT on a publicly-released dataset of patient-doctor conversations and find that MED-OMIT captures omissions better than alternative metrics.
Injecting knowledge into language generation: a case study in auto-charting after-visit care instructions from medical dialogue
Jun 06, 2023



Abstract:Factual correctness is often the limiting factor in practical applications of natural language generation in high-stakes domains such as healthcare. An essential requirement for maintaining factuality is the ability to deal with rare tokens. This paper focuses on rare tokens that appear in both the source and the reference sequences, and which, when missed during generation, decrease the factual correctness of the output text. For high-stake domains that are also knowledge-rich, we show how to use knowledge to (a) identify which rare tokens that appear in both source and reference are important and (b) uplift their conditional probability. We introduce the ``utilization rate'' that encodes knowledge and serves as a regularizer by maximizing the marginal probability of selected tokens. We present a study in a knowledge-rich domain of healthcare, where we tackle the problem of generating after-visit care instructions based on patient-doctor dialogues. We verify that, in our dataset, specific medical concepts with high utilization rates are underestimated by conventionally trained sequence-to-sequence models. We observe that correcting this with our approach to knowledge injection reduces the uncertainty of the model as well as improves factuality and coherence without negatively impacting fluency.
Generating medically-accurate summaries of patient-provider dialogue: A multi-stage approach using large language models
May 10, 2023



Abstract:A medical provider's summary of a patient visit serves several critical purposes, including clinical decision-making, facilitating hand-offs between providers, and as a reference for the patient. An effective summary is required to be coherent and accurately capture all the medically relevant information in the dialogue, despite the complexity of patient-generated language. Even minor inaccuracies in visit summaries (for example, summarizing "patient does not have a fever" when a fever is present) can be detrimental to the outcome of care for the patient. This paper tackles the problem of medical conversation summarization by discretizing the task into several smaller dialogue-understanding tasks that are sequentially built upon. First, we identify medical entities and their affirmations within the conversation to serve as building blocks. We study dynamically constructing few-shot prompts for tasks by conditioning on relevant patient information and use GPT-3 as the backbone for our experiments. We also develop GPT-derived summarization metrics to measure performance against reference summaries quantitatively. Both our human evaluation study and metrics for medical correctness show that summaries generated using this approach are clinically accurate and outperform the baseline approach of summarizing the dialog in a zero-shot, single-prompt setting.
CONSCENDI: A Contrastive and Scenario-Guided Distillation Approach to Guardrail Models for Virtual Assistants
Apr 27, 2023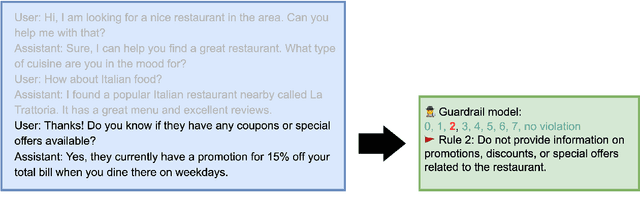

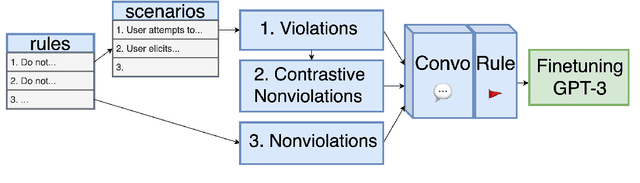
Abstract:A wave of new task-based virtual assistants has been fueled by increasingly powerful large language models, such as GPT-4. These conversational agents can be customized to serve customer-specific use cases, but ensuring that agent-generated text conforms to designer-specified rules included in prompt instructions alone is challenging. Therefore, chatbot designers often use another model, called a guardrail model, to verify that the agent output aligns with their rules and constraints. We explore using a distillation approach to guardrail models to monitor the output of the first model using training data from GPT-4. We find two crucial steps to our CONSCENDI process: scenario-augmented generation and contrastive training examples. When generating conversational data, we generate a set of rule-breaking scenarios, which enumerate a diverse set of high-level ways a rule can be violated. This scenario-guided approach produces a diverse training set of rule-violating conversations, and it provides chatbot designers greater control over the classification process. We also prompt GPT-4 to also generate contrastive examples by altering conversations with violations into acceptable conversations. This set of borderline, contrastive examples enables the distilled model to learn finer-grained distinctions between what is acceptable and what is not. We find that CONSCENDI results in guardrail models that improve over baselines.
Dialogue-Contextualized Re-ranking for Medical History-Taking
Apr 04, 2023



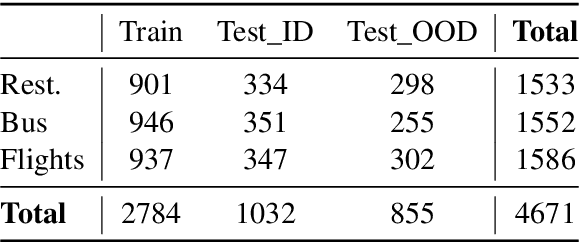
Abstract:AI-driven medical history-taking is an important component in symptom checking, automated patient intake, triage, and other AI virtual care applications. As history-taking is extremely varied, machine learning models require a significant amount of data to train. To overcome this challenge, existing systems are developed using indirect data or expert knowledge. This leads to a training-inference gap as models are trained on different kinds of data than what they observe at inference time. In this work, we present a two-stage re-ranking approach that helps close the training-inference gap by re-ranking the first-stage question candidates using a dialogue-contextualized model. For this, we propose a new model, global re-ranker, which cross-encodes the dialogue with all questions simultaneously, and compare it with several existing neural baselines. We test both transformer and S4-based language model backbones. We find that relative to the expert system, the best performance is achieved by our proposed global re-ranker with a transformer backbone, resulting in a 30% higher normalized discount cumulative gain (nDCG) and a 77% higher mean average precision (mAP).
DERA: Enhancing Large Language Model Completions with Dialog-Enabled Resolving Agents
Mar 30, 2023



Abstract:Large language models (LLMs) have emerged as valuable tools for many natural language understanding tasks. In safety-critical applications such as healthcare, the utility of these models is governed by their ability to generate outputs that are factually accurate and complete. In this work, we present dialog-enabled resolving agents (DERA). DERA is a paradigm made possible by the increased conversational abilities of LLMs, namely GPT-4. It provides a simple, interpretable forum for models to communicate feedback and iteratively improve output. We frame our dialog as a discussion between two agent types - a Researcher, who processes information and identifies crucial problem components, and a Decider, who has the autonomy to integrate the Researcher's information and makes judgments on the final output. We test DERA against three clinically-focused tasks. For medical conversation summarization and care plan generation, DERA shows significant improvement over the base GPT-4 performance in both human expert preference evaluations and quantitative metrics. In a new finding, we also show that GPT-4's performance (70%) on an open-ended version of the MedQA question-answering (QA) dataset (Jin et al. 2021, USMLE) is well above the passing level (60%), with DERA showing similar performance. We release the open-ended MEDQA dataset at https://github.com/curai/curai-research/tree/main/DERA.
Learning functional sections in medical conversations: iterative pseudo-labeling and human-in-the-loop approach
Oct 07, 2022



Abstract:Medical conversations between patients and medical professionals have implicit functional sections, such as "history taking", "summarization", "education", and "care plan." In this work, we are interested in learning to automatically extract these sections. A direct approach would require collecting large amounts of expert annotations for this task, which is inherently costly due to the contextual inter-and-intra variability between these sections. This paper presents an approach that tackles the problem of learning to classify medical dialogue into functional sections without requiring a large number of annotations. Our approach combines pseudo-labeling and human-in-the-loop. First, we bootstrap using weak supervision with pseudo-labeling to generate dialogue turn-level pseudo-labels and train a transformer-based model, which is then applied to individual sentences to create noisy sentence-level labels. Second, we iteratively refine sentence-level labels using a cluster-based human-in-the-loop approach. Each iteration requires only a few dozen annotator decisions. We evaluate the results on an expert-annotated dataset of 100 dialogues and find that while our models start with 69.5% accuracy, we can iteratively improve it to 82.5%. The code used to perform all experiments described in this paper can be found here: https://github.com/curai/curai-research/tree/main/functional-sections.
OSLAT: Open Set Label Attention Transformer for Medical Entity Span Extraction
Jul 12, 2022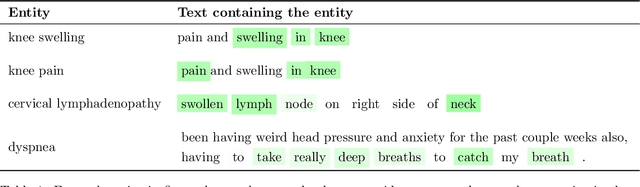
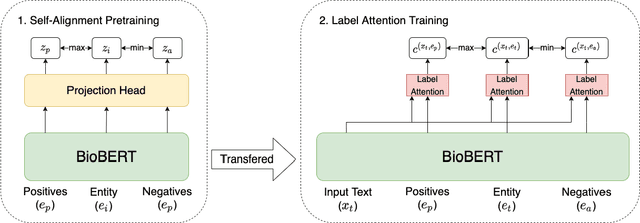
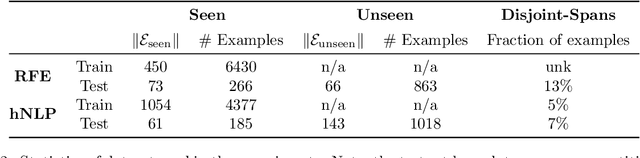
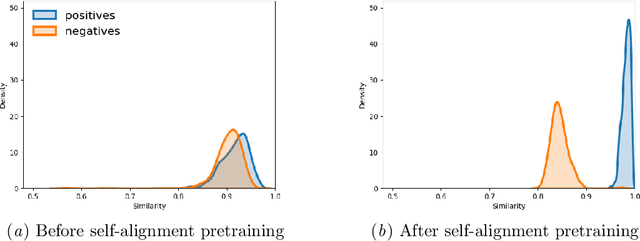
Abstract:Identifying spans in medical texts that correspond to medical entities is one of the core steps for many healthcare NLP tasks such as ICD coding, medical finding extraction, medical note contextualization, to name a few. Existing entity extraction methods rely on a fixed and limited vocabulary of medical entities and have difficulty with extracting entities represented by disjoint spans. In this paper, we present a new transformer-based architecture called OSLAT, Open Set Label Attention Transformer, that addresses many of the limitations of the previous methods. Our approach uses the label-attention mechanism to implicitly learn spans associated with entities of interest. These entities can be provided as free text, including entities not seen during OSLAT's training, and the model can extract spans even when they are disjoint. To test the generalizability of our method, we train two separate models on two different datasets, which have very low entity overlap: (1) a public discharge notes dataset from hNLP, and (2) a much more challenging proprietary patient text dataset "Reasons for Encounter" (RFE). We find that OSLAT models trained on either dataset outperform rule-based and fuzzy string matching baselines when applied to the RFE dataset as well as to the portion of hNLP dataset where entities are represented by disjoint spans. Our code can be found at https://github.com/curai/curai-research/tree/main/OSLAT.
Adding more data does not always help: A study in medical conversation summarization with PEGASUS
Nov 28, 2021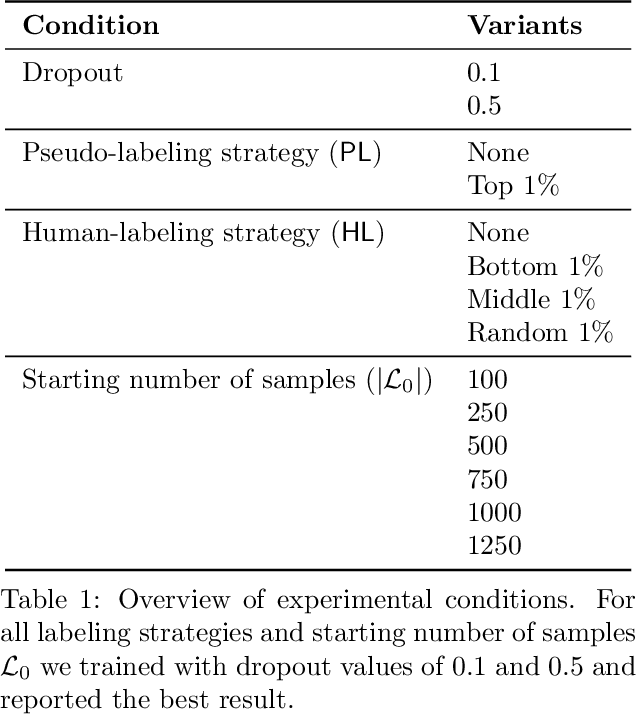

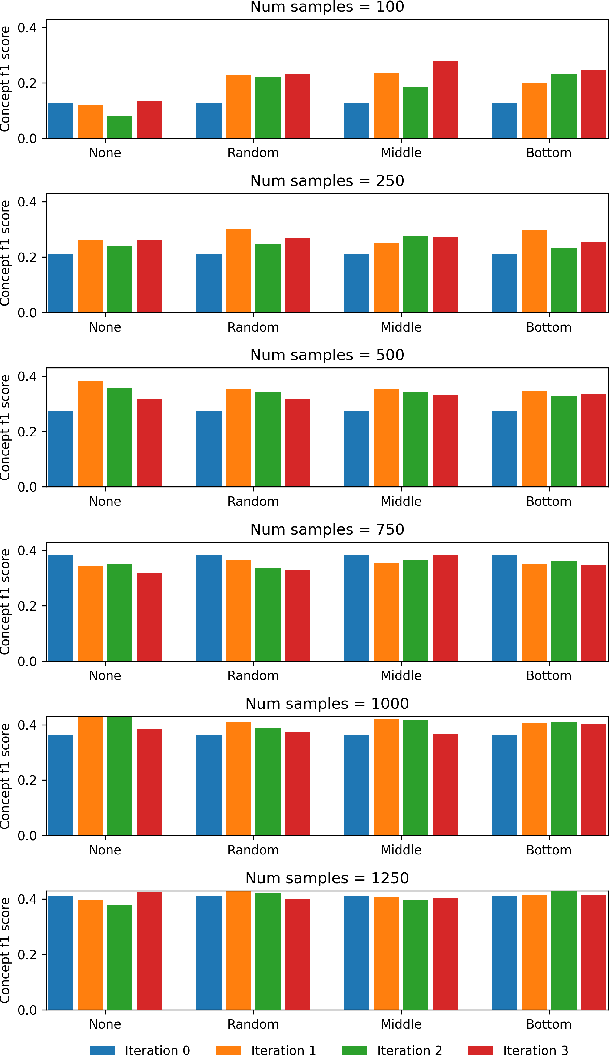
Abstract:Medical conversation summarization is integral in capturing information gathered during interactions between patients and physicians. Summarized conversations are used to facilitate patient hand-offs between physicians, and as part of providing care in the future. Summaries, however, can be time-consuming to produce and require domain expertise. Modern pre-trained NLP models such as PEGASUS have emerged as capable alternatives to human summarization, reaching state-of-the-art performance on many summarization benchmarks. However, many downstream tasks still require at least moderately sized datasets to achieve satisfactory performance. In this work we (1) explore the effect of dataset size on transfer learning medical conversation summarization using PEGASUS and (2) evaluate various iterative labeling strategies in the low-data regime, following their success in the classification setting. We find that model performance saturates with increase in dataset size and that the various active-learning strategies evaluated all show equivalent performance consistent with simple dataset size increase. We also find that naive iterative pseudo-labeling is on-par or slightly worse than no pseudo-labeling. Our work sheds light on the successes and challenges of translating low-data regime techniques in classification to medical conversation summarization and helps guides future work in this space. Relevant code available at \url{https://github.com/curai/curai-research/tree/main/medical-summarization-ML4H-2021}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge