OSLAT: Open Set Label Attention Transformer for Medical Entity Span Extraction
Paper and Code
Jul 12, 2022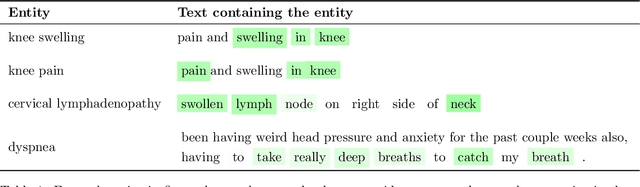
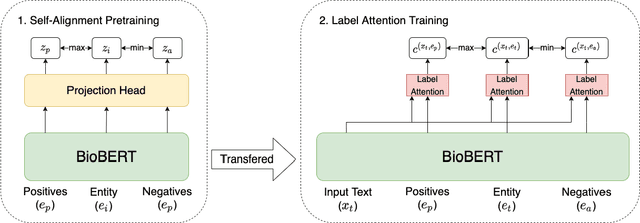
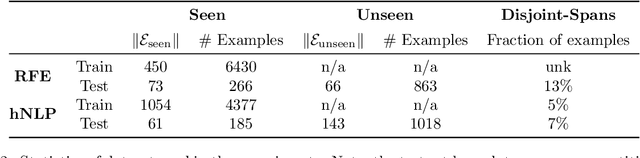
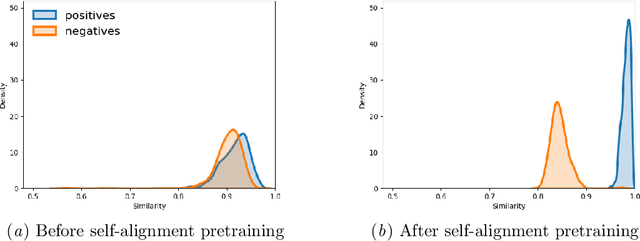
Identifying spans in medical texts that correspond to medical entities is one of the core steps for many healthcare NLP tasks such as ICD coding, medical finding extraction, medical note contextualization, to name a few. Existing entity extraction methods rely on a fixed and limited vocabulary of medical entities and have difficulty with extracting entities represented by disjoint spans. In this paper, we present a new transformer-based architecture called OSLAT, Open Set Label Attention Transformer, that addresses many of the limitations of the previous methods. Our approach uses the label-attention mechanism to implicitly learn spans associated with entities of interest. These entities can be provided as free text, including entities not seen during OSLAT's training, and the model can extract spans even when they are disjoint. To test the generalizability of our method, we train two separate models on two different datasets, which have very low entity overlap: (1) a public discharge notes dataset from hNLP, and (2) a much more challenging proprietary patient text dataset "Reasons for Encounter" (RFE). We find that OSLAT models trained on either dataset outperform rule-based and fuzzy string matching baselines when applied to the RFE dataset as well as to the portion of hNLP dataset where entities are represented by disjoint spans. Our code can be found at https://github.com/curai/curai-research/tree/main/OSLAT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge