Gregory N. McKay
A Deep Learning Bidirectional Temporal Tracking Algorithm for Automated Blood Cell Counting from Non-invasive Capillaroscopy Videos
Dec 09, 2020
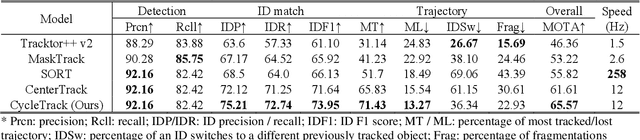
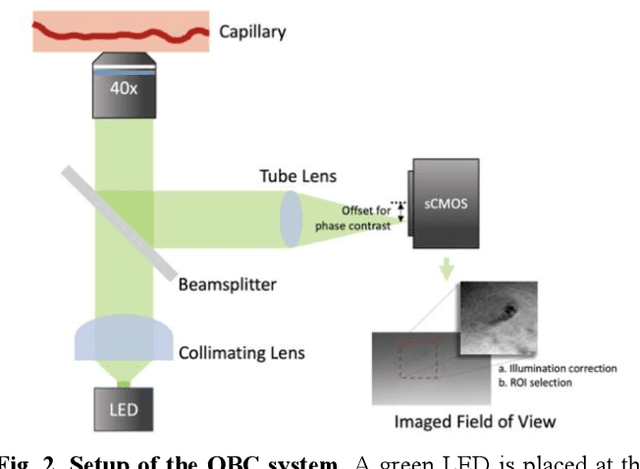

Abstract:Oblique back-illumination capillaroscopy has recently been introduced as a method for high-quality, non-invasive blood cell imaging in human capillaries. To make this technique practical for clinical blood cell counting, solutions for automatic processing of acquired videos are needed. Here, we take the first step towards this goal, by introducing a deep learning multi-cell tracking model, named CycleTrack, which achieves accurate blood cell counting from capillaroscopic videos. CycleTrack combines two simple online tracking models, SORT and CenterTrack, and is tailored to features of capillary blood cell flow. Blood cells are tracked by displacement vectors in two opposing temporal directions (forward- and backward-tracking) between consecutive frames. This approach yields accurate tracking despite rapidly moving and deforming blood cells. The proposed model outperforms other baseline trackers, achieving 65.57% Multiple Object Tracking Accuracy and 73.95% ID F1 score on test videos. Compared to manual blood cell counting, CycleTrack achieves 96.58 $\pm$ 2.43% cell counting accuracy among 8 test videos with 1000 frames each compared to 93.45% and 77.02% accuracy for independent CenterTrack and SORT almost without additional time expense. It takes 800s to track and count approximately 8000 blood cells from 9,600 frames captured in a typical one-minute video. Moreover, the blood cell velocity measured by CycleTrack demonstrates a consistent, pulsatile pattern within the physiological range of heart rate. Lastly, we discuss future improvements for the CycleTrack framework, which would enable clinical translation of the oblique back-illumination microscope towards a real-time and non-invasive point-of-care blood cell counting and analyzing technology.
Deep Adversarial Training for Multi-Organ Nuclei Segmentation in Histopathology Images
Oct 19, 2018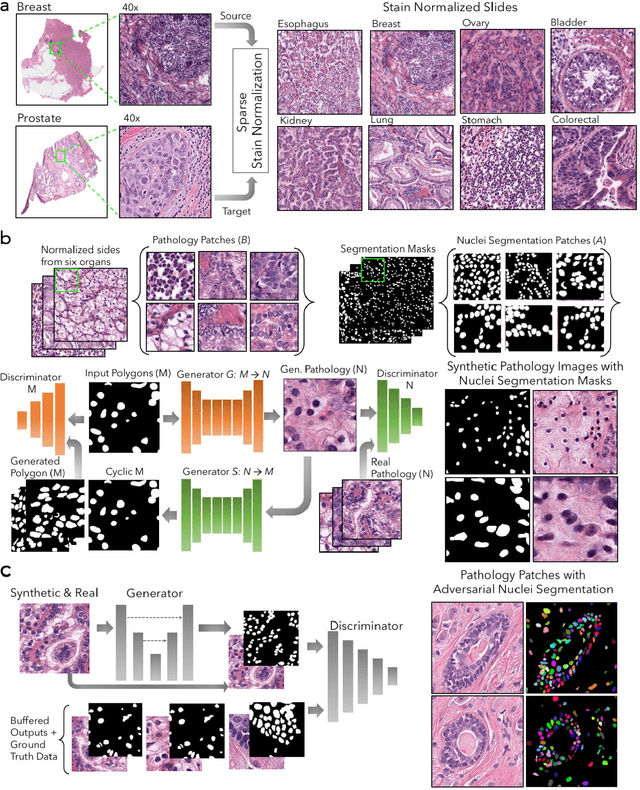
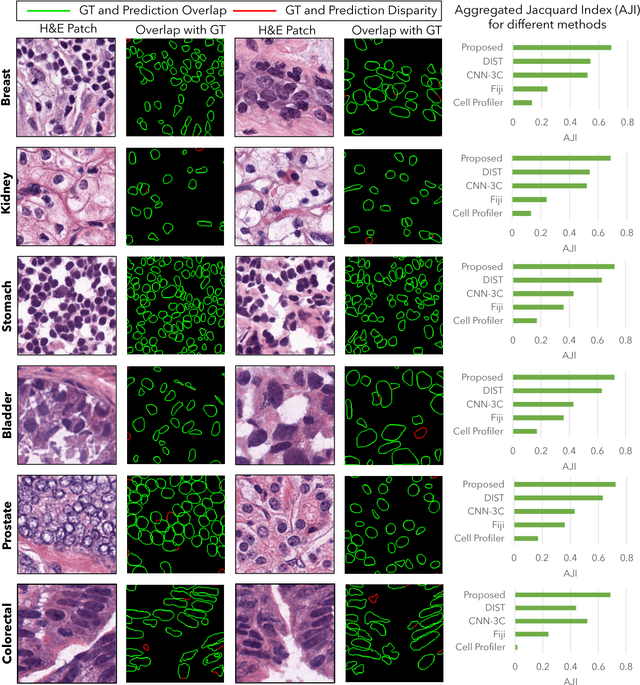
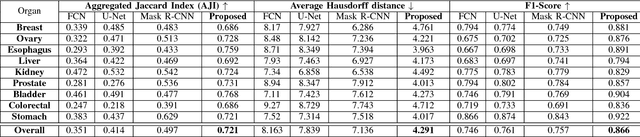
Abstract:Nuclei segmentation is a fundamental task that is critical for various computational pathology applications including nuclei morphology analysis, cell type classification, and cancer grading. Conventional vision-based methods for nuclei segmentation struggle in challenging cases and deep learning approaches have proven to be more robust and generalizable. However, CNNs require large amounts of labeled histopathology data. Moreover, conventional CNN-based approaches lack structured prediction capabilities which are required to distinguish overlapping and clumped nuclei. Here, we present an approach to nuclei segmentation that overcomes these challenges by utilizing a conditional generative adversarial network (cGAN) trained with synthetic and real data. We generate a large dataset of H&E training images with perfect nuclei segmentation labels using an unpaired GAN framework. This synthetic data along with real histopathology data from six different organs are used to train a conditional GAN with spectral normalization and gradient penalty for nuclei segmentation. This adversarial regression framework enforces higher order consistency when compared to conventional CNN models. We demonstrate that this nuclei segmentation approach generalizes across different organs, sites, patients and disease states, and outperforms conventional approaches, especially in isolating individual and overlapping nuclei.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge