Jiaheng Zhang
MemPot: Defending Against Memory Extraction Attack with Optimized Honeypots
Feb 07, 2026Abstract:Large Language Model (LLM)-based agents employ external and internal memory systems to handle complex, goal-oriented tasks, yet this exposes them to severe extraction attacks, and effective defenses remain lacking. In this paper, we propose MemPot, the first theoretically verified defense framework against memory extraction attacks by injecting optimized honeypots into the memory. Through a two-stage optimization process, MemPot generates trap documents that maximize the retrieval probability for attackers while remaining inconspicuous to benign users. We model the detection process as Wald's Sequential Probability Ratio Test (SPRT) and theoretically prove that MemPot achieves a lower average number of sampling rounds compared to optimal static detectors. Empirically, MemPot significantly outperforms state-of-the-art baselines, achieving a 50% improvement in detection AUROC and an 80% increase in True Positive Rate under low False Positive Rate constraints. Furthermore, our experiments confirm that MemPot incurs zero additional online inference latency and preserves the agent's utility on standard tasks, verifying its superiority in safety, harmlessness, and efficiency.
Secure Code Generation via Online Reinforcement Learning with Vulnerability Reward Model
Feb 07, 2026Abstract:Large language models (LLMs) are increasingly used in software development, yet their tendency to generate insecure code remains a major barrier to real-world deployment. Existing secure code alignment methods often suffer from a functionality--security paradox, improving security at the cost of substantial utility degradation. We propose SecCoderX, an online reinforcement learning framework for functionality-preserving secure code generation. SecCoderX first bridges vulnerability detection and secure code generation by repurposing mature detection resources in two ways: (i) synthesizing diverse, reality-grounded vulnerability-inducing coding tasks for online RL rollouts, and (ii) training a reasoning-based vulnerability reward model that provides scalable and reliable security supervision. Together, these components are unified in an online RL loop to align code LLMs to generate secure and functional code. Extensive experiments demonstrate that SecCoderX achieves state-of-the-art performance, improving Effective Safety Rate (ESR) by approximately 10% over unaligned models, whereas prior methods often degrade ESR by 14-54%. We release our code, dataset and model checkpoints at https://github.com/AndrewWTY/SecCoderX.
Reliable and Responsible Foundation Models: A Comprehensive Survey
Feb 04, 2026Abstract:Foundation models, including Large Language Models (LLMs), Multimodal Large Language Models (MLLMs), Image Generative Models (i.e, Text-to-Image Models and Image-Editing Models), and Video Generative Models, have become essential tools with broad applications across various domains such as law, medicine, education, finance, science, and beyond. As these models see increasing real-world deployment, ensuring their reliability and responsibility has become critical for academia, industry, and government. This survey addresses the reliable and responsible development of foundation models. We explore critical issues, including bias and fairness, security and privacy, uncertainty, explainability, and distribution shift. Our research also covers model limitations, such as hallucinations, as well as methods like alignment and Artificial Intelligence-Generated Content (AIGC) detection. For each area, we review the current state of the field and outline concrete future research directions. Additionally, we discuss the intersections between these areas, highlighting their connections and shared challenges. We hope our survey fosters the development of foundation models that are not only powerful but also ethical, trustworthy, reliable, and socially responsible.
Beyond Hard Masks: Progressive Token Evolution for Diffusion Language Models
Jan 12, 2026Abstract:Diffusion Language Models (DLMs) offer a promising alternative for language modeling by enabling parallel decoding through iterative refinement. However, most DLMs rely on hard binary masking and discrete token assignments, which hinder the revision of early decisions and underutilize intermediate probabilistic representations. In this paper, we propose EvoToken-DLM, a novel diffusion-based language modeling approach that replaces hard binary masks with evolving soft token distributions. EvoToken-DLM enables a progressive transition from masked states to discrete outputs, supporting revisable decoding. To effectively support this evolution, we introduce continuous trajectory supervision, which aligns training objectives with iterative probabilistic updates. Extensive experiments across multiple benchmarks show that EvoToken-DLM consistently achieves superior performance, outperforming strong diffusion-based and masked DLM baselines. Project webpage: https://aim-uofa.github.io/EvoTokenDLM.
Reasoning with Confidence: Efficient Verification of LLM Reasoning Steps via Uncertainty Heads
Nov 11, 2025Abstract:Solving complex tasks usually requires LLMs to generate long multi-step reasoning chains. Previous work has shown that verifying the correctness of individual reasoning steps can further improve the performance and efficiency of LLMs on such tasks and enhance solution interpretability. However, existing verification approaches, such as Process Reward Models (PRMs), are either computationally expensive, limited to specific domains, or require large-scale human or model-generated annotations. Thus, we propose a lightweight alternative for step-level reasoning verification based on data-driven uncertainty scores. We train transformer-based uncertainty quantification heads (UHeads) that use the internal states of a frozen LLM to estimate the uncertainty of its reasoning steps during generation. The approach is fully automatic: target labels are generated either by another larger LLM (e.g., DeepSeek R1) or in a self-supervised manner by the original model itself. UHeads are both effective and lightweight, containing less than 10M parameters. Across multiple domains, including mathematics, planning, and general knowledge question answering, they match or even surpass the performance of PRMs that are up to 810x larger. Our findings suggest that the internal states of LLMs encode their uncertainty and can serve as reliable signals for reasoning verification, offering a promising direction toward scalable and generalizable introspective LLMs.
SWAP: Towards Copyright Auditing of Soft Prompts via Sequential Watermarking
Nov 05, 2025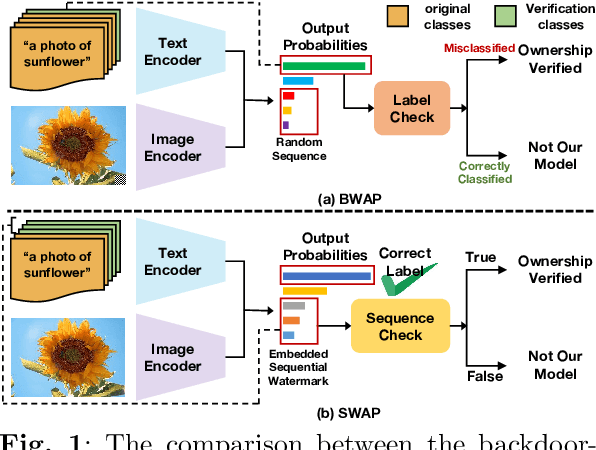
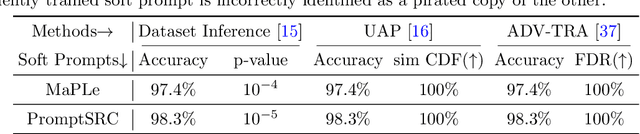
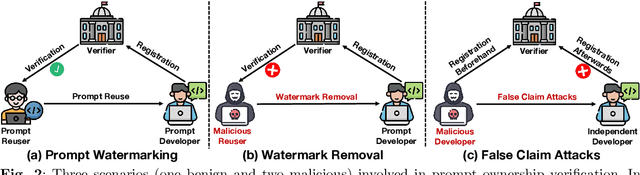
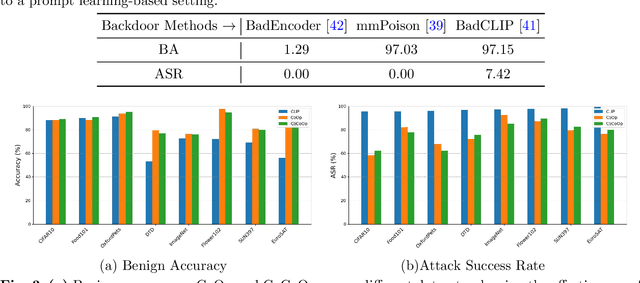
Abstract:Large-scale vision-language models, especially CLIP, have demonstrated remarkable performance across diverse downstream tasks. Soft prompts, as carefully crafted modules that efficiently adapt vision-language models to specific tasks, necessitate effective copyright protection. In this paper, we investigate model copyright protection by auditing whether suspicious third-party models incorporate protected soft prompts. While this can be viewed as a special case of model ownership auditing, our analysis shows that existing techniques are ineffective due to prompt learning's unique characteristics. Non-intrusive auditing is inherently prone to false positives when independent models share similar data distributions with victim models. Intrusive approaches also fail: backdoor methods designed for CLIP cannot embed functional triggers, while extending traditional DNN backdoor techniques to prompt learning suffers from harmfulness and ambiguity challenges. We find that these failures in intrusive auditing stem from the same fundamental reason: watermarking operates within the same decision space as the primary task yet pursues opposing objectives. Motivated by these findings, we propose sequential watermarking for soft prompts (SWAP), which implants watermarks into a different and more complex space. SWAP encodes watermarks through a specific order of defender-specified out-of-distribution classes, inspired by the zero-shot prediction capability of CLIP. This watermark, which is embedded in a more complex space, keeps the original prediction label unchanged, making it less opposed to the primary task. We further design a hypothesis-test-guided verification protocol for SWAP and provide theoretical analyses of success conditions. Extensive experiments on 11 datasets demonstrate SWAP's effectiveness, harmlessness, and robustness against potential adaptive attacks.
TrojanTO: Action-Level Backdoor Attacks against Trajectory Optimization Models
Jun 15, 2025Abstract:Recent advances in Trajectory Optimization (TO) models have achieved remarkable success in offline reinforcement learning. However, their vulnerabilities against backdoor attacks are poorly understood. We find that existing backdoor attacks in reinforcement learning are based on reward manipulation, which are largely ineffective against the TO model due to its inherent sequence modeling nature. Moreover, the complexities introduced by high-dimensional action spaces further compound the challenge of action manipulation. To address these gaps, we propose TrojanTO, the first action-level backdoor attack against TO models. TrojanTO employs alternating training to enhance the connection between triggers and target actions for attack effectiveness. To improve attack stealth, it utilizes precise poisoning via trajectory filtering for normal performance and batch poisoning for trigger consistency. Extensive evaluations demonstrate that TrojanTO effectively implants backdoor attacks across diverse tasks and attack objectives with a low attack budget (0.3\% of trajectories). Furthermore, TrojanTO exhibits broad applicability to DT, GDT, and DC, underscoring its scalability across diverse TO model architectures.
Efficient Reasoning via Chain of Unconscious Thought
May 26, 2025Abstract:Large Reasoning Models (LRMs) achieve promising performance but compromise token efficiency due to verbose reasoning processes. Unconscious Thought Theory (UTT) posits that complex problems can be solved more efficiently through internalized cognitive processes. Inspired by UTT, we propose a new reasoning paradigm, termed Chain of Unconscious Thought (CoUT), to improve the token efficiency of LRMs by guiding them to mimic human unconscious thought and internalize reasoning processes. Concretely, we first prompt the model to internalize the reasoning by thinking in the hidden layer. Then, we design a bag of token-efficient strategies to further help models reduce unnecessary tokens yet preserve the performance. Our work reveals that models may possess beneficial unconscious thought, enabling improved efficiency without sacrificing performance. Extensive experiments demonstrate the effectiveness of CoUT. Remarkably, it surpasses CoT by reducing token usage by 47.62% while maintaining comparable accuracy, as shown in Figure 1. The code of CoUT is available at this link: https://github.com/Rohan-GRH/CoUT
Silent Leaks: Implicit Knowledge Extraction Attack on RAG Systems through Benign Queries
May 21, 2025Abstract:Retrieval-Augmented Generation (RAG) systems enhance large language models (LLMs) by incorporating external knowledge bases, but they are vulnerable to privacy risks from data extraction attacks. Existing extraction methods typically rely on malicious inputs such as prompt injection or jailbreaking, making them easily detectable via input- or output-level detection. In this paper, we introduce Implicit Knowledge Extraction Attack (IKEA), which conducts knowledge extraction on RAG systems through benign queries. IKEA first leverages anchor concepts to generate queries with the natural appearance, and then designs two mechanisms to lead to anchor concept thoroughly 'explore' the RAG's privacy knowledge: (1) Experience Reflection Sampling, which samples anchor concepts based on past query-response patterns to ensure the queries' relevance to RAG documents; (2) Trust Region Directed Mutation, which iteratively mutates anchor concepts under similarity constraints to further exploit the embedding space. Extensive experiments demonstrate IKEA's effectiveness under various defenses, surpassing baselines by over 80% in extraction efficiency and 90% in attack success rate. Moreover, the substitute RAG system built from IKEA's extractions consistently outperforms those based on baseline methods across multiple evaluation tasks, underscoring the significant privacy risk in RAG systems.
GuardReasoner-VL: Safeguarding VLMs via Reinforced Reasoning
May 16, 2025Abstract:To enhance the safety of VLMs, this paper introduces a novel reasoning-based VLM guard model dubbed GuardReasoner-VL. The core idea is to incentivize the guard model to deliberatively reason before making moderation decisions via online RL. First, we construct GuardReasoner-VLTrain, a reasoning corpus with 123K samples and 631K reasoning steps, spanning text, image, and text-image inputs. Then, based on it, we cold-start our model's reasoning ability via SFT. In addition, we further enhance reasoning regarding moderation through online RL. Concretely, to enhance diversity and difficulty of samples, we conduct rejection sampling followed by data augmentation via the proposed safety-aware data concatenation. Besides, we use a dynamic clipping parameter to encourage exploration in early stages and exploitation in later stages. To balance performance and token efficiency, we design a length-aware safety reward that integrates accuracy, format, and token cost. Extensive experiments demonstrate the superiority of our model. Remarkably, it surpasses the runner-up by 19.27% F1 score on average. We release data, code, and models (3B/7B) of GuardReasoner-VL at https://github.com/yueliu1999/GuardReasoner-VL/
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge