Caleb Geniesse
UC Berkeley/LBNL/ICSI
Landscaper: Understanding Loss Landscapes Through Multi-Dimensional Topological Analysis
Feb 06, 2026Abstract:Loss landscapes are a powerful tool for understanding neural network optimization and generalization, yet traditional low-dimensional analyses often miss complex topological features. We present Landscaper, an open-source Python package for arbitrary-dimensional loss landscape analysis. Landscaper combines Hessian-based subspace construction with topological data analysis to reveal geometric structures such as basin hierarchy and connectivity. A key component is the Saddle-Minimum Average Distance (SMAD) for quantifying landscape smoothness. We demonstrate Landscaper's effectiveness across various architectures and tasks, including those involving pre-trained language models, showing that SMAD captures training transitions, such as landscape simplification, that conventional metrics miss. We also illustrate Landscaper's performance in challenging chemical property prediction tasks, where SMAD can serve as a metric for out-of-distribution generalization, offering valuable insights for model diagnostics and architecture design in data-scarce scientific machine learning scenarios.
Removing Watermarks with Partial Regeneration using Semantic Information
May 13, 2025Abstract:As AI-generated imagery becomes ubiquitous, invisible watermarks have emerged as a primary line of defense for copyright and provenance. The newest watermarking schemes embed semantic signals - content-aware patterns that are designed to survive common image manipulations - yet their true robustness against adaptive adversaries remains under-explored. We expose a previously unreported vulnerability and introduce SemanticRegen, a three-stage, label-free attack that erases state-of-the-art semantic and invisible watermarks while leaving an image's apparent meaning intact. Our pipeline (i) uses a vision-language model to obtain fine-grained captions, (ii) extracts foreground masks with zero-shot segmentation, and (iii) inpaints only the background via an LLM-guided diffusion model, thereby preserving salient objects and style cues. Evaluated on 1,000 prompts across four watermarking systems - TreeRing, StegaStamp, StableSig, and DWT/DCT - SemanticRegen is the only method to defeat the semantic TreeRing watermark (p = 0.10 > 0.05) and reduces bit-accuracy below 0.75 for the remaining schemes, all while maintaining high perceptual quality (masked SSIM = 0.94 +/- 0.01). We further introduce masked SSIM (mSSIM) to quantify fidelity within foreground regions, showing that our attack achieves up to 12 percent higher mSSIM than prior diffusion-based attackers. These results highlight an urgent gap between current watermark defenses and the capabilities of adaptive, semantics-aware adversaries, underscoring the need for watermarking algorithms that are resilient to content-preserving regenerative attacks.
Loss Landscape Analysis for Reliable Quantized ML Models for Scientific Sensing
Feb 12, 2025Abstract:In this paper, we propose a method to perform empirical analysis of the loss landscape of machine learning (ML) models. The method is applied to two ML models for scientific sensing, which necessitates quantization to be deployed and are subject to noise and perturbations due to experimental conditions. Our method allows assessing the robustness of ML models to such effects as a function of quantization precision and under different regularization techniques -- two crucial concerns that remained underexplored so far. By investigating the interplay between performance, efficiency, and robustness by means of loss landscape analysis, we both established a strong correlation between gently-shaped landscapes and robustness to input and weight perturbations and observed other intriguing and non-obvious phenomena. Our method allows a systematic exploration of such trade-offs a priori, i.e., without training and testing multiple models, leading to more efficient development workflows. This work also highlights the importance of incorporating robustness into the Pareto optimization of ML models, enabling more reliable and adaptive scientific sensing systems.
LossLens: Diagnostics for Machine Learning through Loss Landscape Visual Analytics
Dec 17, 2024



Abstract:Modern machine learning often relies on optimizing a neural network's parameters using a loss function to learn complex features. Beyond training, examining the loss function with respect to a network's parameters (i.e., as a loss landscape) can reveal insights into the architecture and learning process. While the local structure of the loss landscape surrounding an individual solution can be characterized using a variety of approaches, the global structure of a loss landscape, which includes potentially many local minima corresponding to different solutions, remains far more difficult to conceptualize and visualize. To address this difficulty, we introduce LossLens, a visual analytics framework that explores loss landscapes at multiple scales. LossLens integrates metrics from global and local scales into a comprehensive visual representation, enhancing model diagnostics. We demonstrate LossLens through two case studies: visualizing how residual connections influence a ResNet-20, and visualizing how physical parameters influence a physics-informed neural network (PINN) solving a simple convection problem.
Visualizing Loss Functions as Topological Landscape Profiles
Nov 19, 2024



Abstract:In machine learning, a loss function measures the difference between model predictions and ground-truth (or target) values. For neural network models, visualizing how this loss changes as model parameters are varied can provide insights into the local structure of the so-called loss landscape (e.g., smoothness) as well as global properties of the underlying model (e.g., generalization performance). While various methods for visualizing the loss landscape have been proposed, many approaches limit sampling to just one or two directions, ignoring potentially relevant information in this extremely high-dimensional space. This paper introduces a new representation based on topological data analysis that enables the visualization of higher-dimensional loss landscapes. After describing this new topological landscape profile representation, we show how the shape of loss landscapes can reveal new details about model performance and learning dynamics, highlighting several use cases, including image segmentation (e.g., UNet) and scientific machine learning (e.g., physics-informed neural networks). Through these examples, we provide new insights into how loss landscapes vary across distinct hyperparameter spaces: we find that the topology of the loss landscape is simpler for better-performing models; and we observe greater variation in the shape of loss landscapes near transitions from low to high model performance.
Evaluating Loss Landscapes from a Topology Perspective
Nov 14, 2024



Abstract:Characterizing the loss of a neural network with respect to model parameters, i.e., the loss landscape, can provide valuable insights into properties of that model. Various methods for visualizing loss landscapes have been proposed, but less emphasis has been placed on quantifying and extracting actionable and reproducible insights from these complex representations. Inspired by powerful tools from topological data analysis (TDA) for summarizing the structure of high-dimensional data, here we characterize the underlying shape (or topology) of loss landscapes, quantifying the topology to reveal new insights about neural networks. To relate our findings to the machine learning (ML) literature, we compute simple performance metrics (e.g., accuracy, error), and we characterize the local structure of loss landscapes using Hessian-based metrics (e.g., largest eigenvalue, trace, eigenvalue spectral density). Following this approach, we study established models from image pattern recognition (e.g., ResNets) and scientific ML (e.g., physics-informed neural networks), and we show how quantifying the shape of loss landscapes can provide new insights into model performance and learning dynamics.
Mitigating Memorization In Language Models
Oct 03, 2024



Abstract:Language models (LMs) can "memorize" information, i.e., encode training data in their weights in such a way that inference-time queries can lead to verbatim regurgitation of that data. This ability to extract training data can be problematic, for example, when data are private or sensitive. In this work, we investigate methods to mitigate memorization: three regularizer-based, three finetuning-based, and eleven machine unlearning-based methods, with five of the latter being new methods that we introduce. We also introduce TinyMem, a suite of small, computationally-efficient LMs for the rapid development and evaluation of memorization-mitigation methods. We demonstrate that the mitigation methods that we develop using TinyMem can successfully be applied to production-grade LMs, and we determine via experiment that: regularizer-based mitigation methods are slow and ineffective at curbing memorization; fine-tuning-based methods are effective at curbing memorization, but overly expensive, especially for retaining higher accuracies; and unlearning-based methods are faster and more effective, allowing for the precise localization and removal of memorized information from LM weights prior to inference. We show, in particular, that our proposed unlearning method BalancedSubnet outperforms other mitigation methods at removing memorized information while preserving performance on target tasks.
Reliable edge machine learning hardware for scientific applications
Jun 27, 2024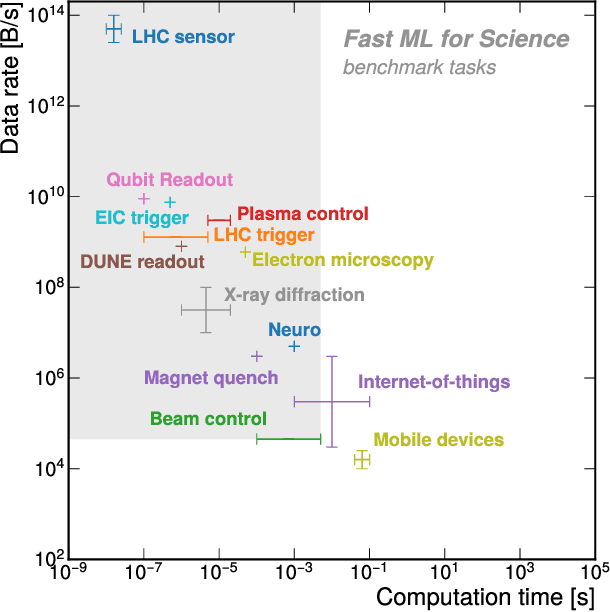
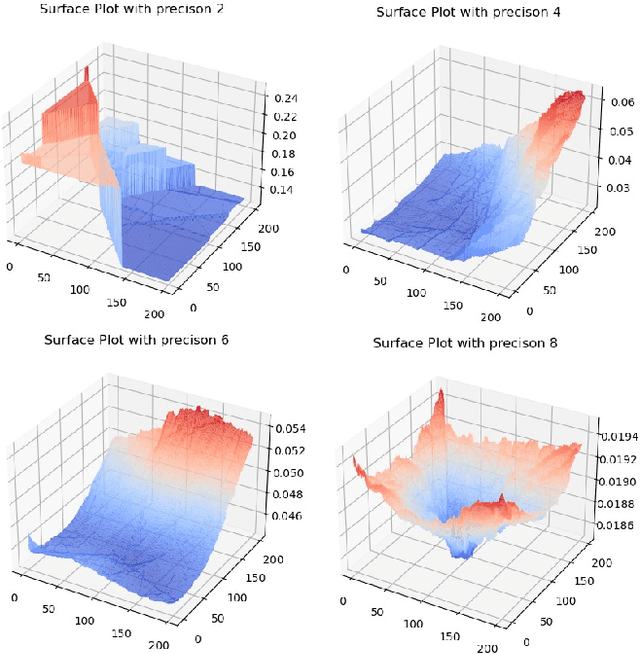
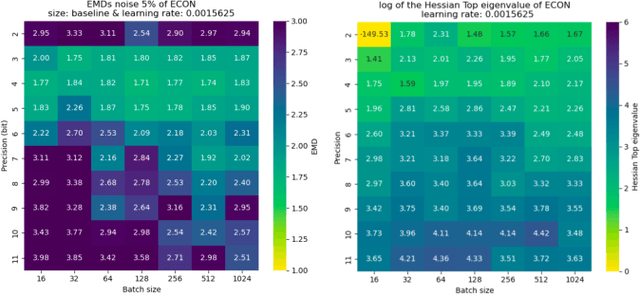
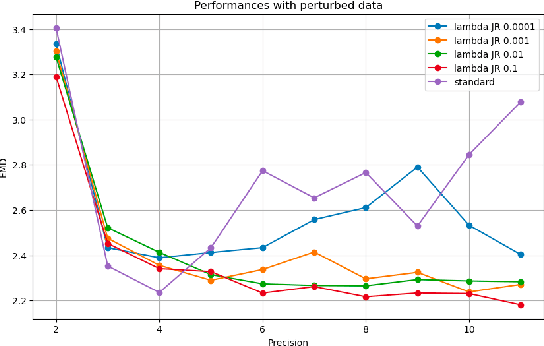
Abstract:Extreme data rate scientific experiments create massive amounts of data that require efficient ML edge processing. This leads to unique validation challenges for VLSI implementations of ML algorithms: enabling bit-accurate functional simulations for performance validation in experimental software frameworks, verifying those ML models are robust under extreme quantization and pruning, and enabling ultra-fine-grained model inspection for efficient fault tolerance. We discuss approaches to developing and validating reliable algorithms at the scientific edge under such strict latency, resource, power, and area requirements in extreme experimental environments. We study metrics for developing robust algorithms, present preliminary results and mitigation strategies, and conclude with an outlook of these and future directions of research towards the longer-term goal of developing autonomous scientific experimentation methods for accelerated scientific discovery.
MoleculeNet: A Benchmark for Molecular Machine Learning
Oct 26, 2018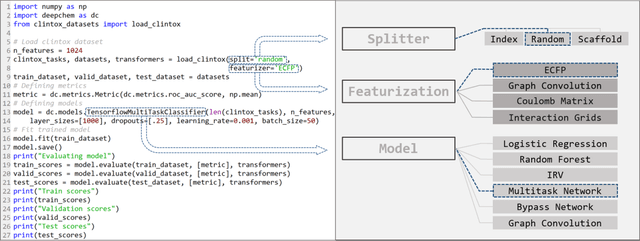
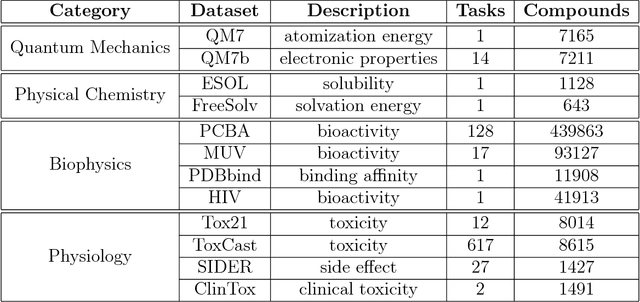
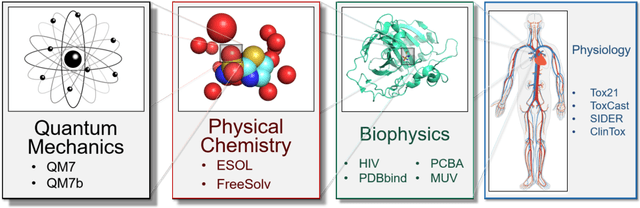
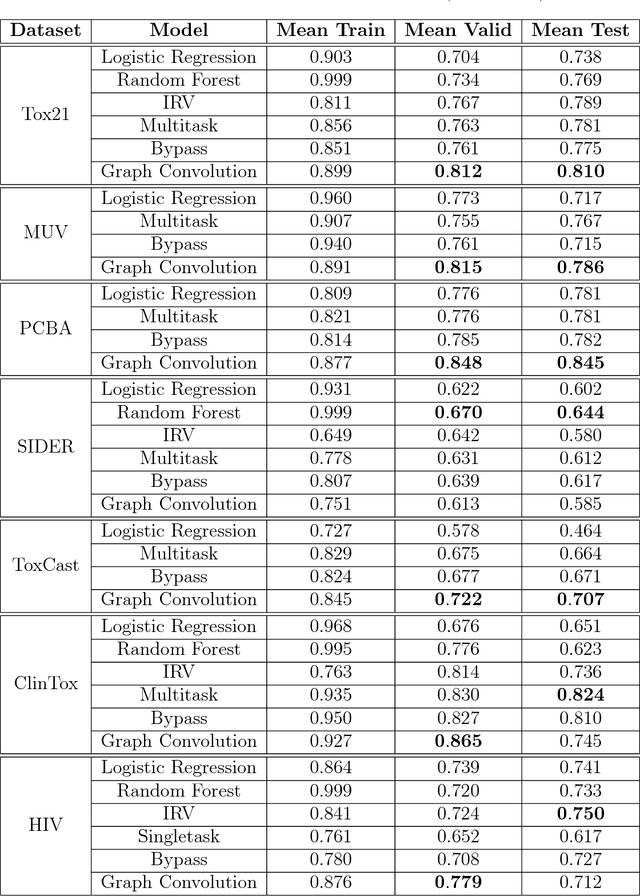
Abstract:Molecular machine learning has been maturing rapidly over the last few years. Improved methods and the presence of larger datasets have enabled machine learning algorithms to make increasingly accurate predictions about molecular properties. However, algorithmic progress has been limited due to the lack of a standard benchmark to compare the efficacy of proposed methods; most new algorithms are benchmarked on different datasets making it challenging to gauge the quality of proposed methods. This work introduces MoleculeNet, a large scale benchmark for molecular machine learning. MoleculeNet curates multiple public datasets, establishes metrics for evaluation, and offers high quality open-source implementations of multiple previously proposed molecular featurization and learning algorithms (released as part of the DeepChem open source library). MoleculeNet benchmarks demonstrate that learnable representations are powerful tools for molecular machine learning and broadly offer the best performance. However, this result comes with caveats. Learnable representations still struggle to deal with complex tasks under data scarcity and highly imbalanced classification. For quantum mechanical and biophysical datasets, the use of physics-aware featurizations can be more important than choice of particular learning algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge