John Kevin Cava
Removing Watermarks with Partial Regeneration using Semantic Information
May 13, 2025Abstract:As AI-generated imagery becomes ubiquitous, invisible watermarks have emerged as a primary line of defense for copyright and provenance. The newest watermarking schemes embed semantic signals - content-aware patterns that are designed to survive common image manipulations - yet their true robustness against adaptive adversaries remains under-explored. We expose a previously unreported vulnerability and introduce SemanticRegen, a three-stage, label-free attack that erases state-of-the-art semantic and invisible watermarks while leaving an image's apparent meaning intact. Our pipeline (i) uses a vision-language model to obtain fine-grained captions, (ii) extracts foreground masks with zero-shot segmentation, and (iii) inpaints only the background via an LLM-guided diffusion model, thereby preserving salient objects and style cues. Evaluated on 1,000 prompts across four watermarking systems - TreeRing, StegaStamp, StableSig, and DWT/DCT - SemanticRegen is the only method to defeat the semantic TreeRing watermark (p = 0.10 > 0.05) and reduces bit-accuracy below 0.75 for the remaining schemes, all while maintaining high perceptual quality (masked SSIM = 0.94 +/- 0.01). We further introduce masked SSIM (mSSIM) to quantify fidelity within foreground regions, showing that our attack achieves up to 12 percent higher mSSIM than prior diffusion-based attackers. These results highlight an urgent gap between current watermark defenses and the capabilities of adaptive, semantics-aware adversaries, underscoring the need for watermarking algorithms that are resilient to content-preserving regenerative attacks.
LossLens: Diagnostics for Machine Learning through Loss Landscape Visual Analytics
Dec 17, 2024



Abstract:Modern machine learning often relies on optimizing a neural network's parameters using a loss function to learn complex features. Beyond training, examining the loss function with respect to a network's parameters (i.e., as a loss landscape) can reveal insights into the architecture and learning process. While the local structure of the loss landscape surrounding an individual solution can be characterized using a variety of approaches, the global structure of a loss landscape, which includes potentially many local minima corresponding to different solutions, remains far more difficult to conceptualize and visualize. To address this difficulty, we introduce LossLens, a visual analytics framework that explores loss landscapes at multiple scales. LossLens integrates metrics from global and local scales into a comprehensive visual representation, enhancing model diagnostics. We demonstrate LossLens through two case studies: visualizing how residual connections influence a ResNet-20, and visualizing how physical parameters influence a physics-informed neural network (PINN) solving a simple convection problem.
AugLoss: A Learning Methodology for Real-World Dataset Corruption
Jun 05, 2022
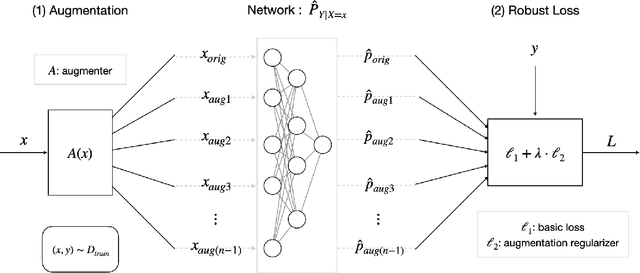
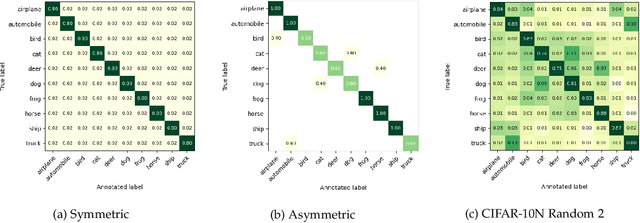
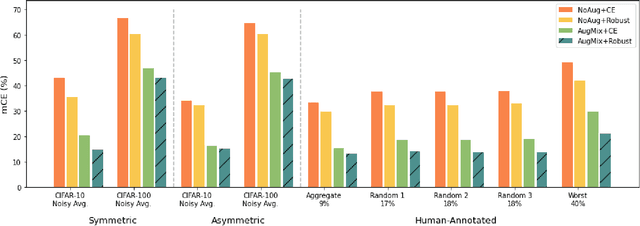
Abstract:Deep Learning (DL) models achieve great successes in many domains. However, DL models increasingly face safety and robustness concerns, including noisy labeling in the training stage and feature distribution shifts in the testing stage. Previous works made significant progress in addressing these problems, but the focus has largely been on developing solutions for only one problem at a time. For example, recent work has argued for the use of tunable robust loss functions to mitigate label noise, and data augmentation (e.g., AugMix) to combat distribution shifts. As a step towards addressing both problems simultaneously, we introduce AugLoss, a simple but effective methodology that achieves robustness against both train-time noisy labeling and test-time feature distribution shifts by unifying data augmentation and robust loss functions. We conduct comprehensive experiments in varied settings of real-world dataset corruption to showcase the gains achieved by AugLoss compared to previous state-of-the-art methods. Lastly, we hope this work will open new directions for designing more robust and reliable DL models under real-world corruptions.
Towards Conditional Generation of Minimal Action Potential Pathways for Molecular Dynamics
Nov 28, 2021

Abstract:In this paper, we utilized generative models, and reformulate it for problems in molecular dynamics (MD) simulation, by introducing an MD potential energy component to our generative model. By incorporating potential energy as calculated from TorchMD into a conditional generative framework, we attempt to construct a low-potential energy route of transformation between the helix~$\rightarrow$~coil structures of a protein. We show how to add an additional loss function to conditional generative models, motivated by potential energy of molecular configurations, and also present an optimization technique for such an augmented loss function. Our results show the benefit of this additional loss term on synthesizing realistic molecular trajectories.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge