Dmitriy Morozov
Visualizing Loss Functions as Topological Landscape Profiles
Nov 19, 2024



Abstract:In machine learning, a loss function measures the difference between model predictions and ground-truth (or target) values. For neural network models, visualizing how this loss changes as model parameters are varied can provide insights into the local structure of the so-called loss landscape (e.g., smoothness) as well as global properties of the underlying model (e.g., generalization performance). While various methods for visualizing the loss landscape have been proposed, many approaches limit sampling to just one or two directions, ignoring potentially relevant information in this extremely high-dimensional space. This paper introduces a new representation based on topological data analysis that enables the visualization of higher-dimensional loss landscapes. After describing this new topological landscape profile representation, we show how the shape of loss landscapes can reveal new details about model performance and learning dynamics, highlighting several use cases, including image segmentation (e.g., UNet) and scientific machine learning (e.g., physics-informed neural networks). Through these examples, we provide new insights into how loss landscapes vary across distinct hyperparameter spaces: we find that the topology of the loss landscape is simpler for better-performing models; and we observe greater variation in the shape of loss landscapes near transitions from low to high model performance.
Evaluating Loss Landscapes from a Topology Perspective
Nov 14, 2024



Abstract:Characterizing the loss of a neural network with respect to model parameters, i.e., the loss landscape, can provide valuable insights into properties of that model. Various methods for visualizing loss landscapes have been proposed, but less emphasis has been placed on quantifying and extracting actionable and reproducible insights from these complex representations. Inspired by powerful tools from topological data analysis (TDA) for summarizing the structure of high-dimensional data, here we characterize the underlying shape (or topology) of loss landscapes, quantifying the topology to reveal new insights about neural networks. To relate our findings to the machine learning (ML) literature, we compute simple performance metrics (e.g., accuracy, error), and we characterize the local structure of loss landscapes using Hessian-based metrics (e.g., largest eigenvalue, trace, eigenvalue spectral density). Following this approach, we study established models from image pattern recognition (e.g., ResNets) and scientific ML (e.g., physics-informed neural networks), and we show how quantifying the shape of loss landscapes can provide new insights into model performance and learning dynamics.
Learning Physics for Unveiling Hidden Earthquake Ground Motions via Conditional Generative Modeling
Jul 21, 2024



Abstract:Predicting high-fidelity ground motions for future earthquakes is crucial for seismic hazard assessment and infrastructure resilience. Conventional empirical simulations suffer from sparse sensor distribution and geographically localized earthquake locations, while physics-based methods are computationally intensive and require accurate representations of Earth structures and earthquake sources. We propose a novel artificial intelligence (AI) simulator, Conditional Generative Modeling for Ground Motion (CGM-GM), to synthesize high-frequency and spatially continuous earthquake ground motion waveforms. CGM-GM leverages earthquake magnitudes and geographic coordinates of earthquakes and sensors as inputs, learning complex wave physics and Earth heterogeneities, without explicit physics constraints. This is achieved through a probabilistic autoencoder that captures latent distributions in the time-frequency domain and variational sequential models for prior and posterior distributions. We evaluate the performance of CGM-GM using small-magnitude earthquake records from the San Francisco Bay Area, a region with high seismic risks. CGM-GM demonstrates a strong potential for outperforming a state-of-the-art non-ergodic empirical ground motion model and shows great promise in seismology and beyond.
Data-Efficient Operator Learning via Unsupervised Pretraining and In-Context Learning
Feb 24, 2024



Abstract:Recent years have witnessed the promise of coupling machine learning methods and physical domain-specific insight for solving scientific problems based on partial differential equations (PDEs). However, being data-intensive, these methods still require a large amount of PDE data. This reintroduces the need for expensive numerical PDE solutions, partially undermining the original goal of avoiding these expensive simulations. In this work, seeking data efficiency, we design unsupervised pretraining and in-context learning methods for PDE operator learning. To reduce the need for training data with simulated solutions, we pretrain neural operators on unlabeled PDE data using reconstruction-based proxy tasks. To improve out-of-distribution performance, we further assist neural operators in flexibly leveraging in-context learning methods, without incurring extra training costs or designs. Extensive empirical evaluations on a diverse set of PDEs demonstrate that our method is highly data-efficient, more generalizable, and even outperforms conventional vision-pretrained models.
Robustifying State-space Models for Long Sequences via Approximate Diagonalization
Oct 02, 2023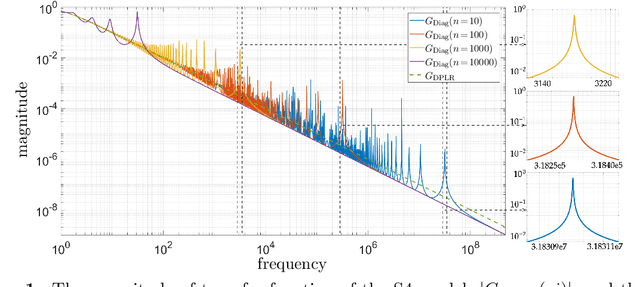
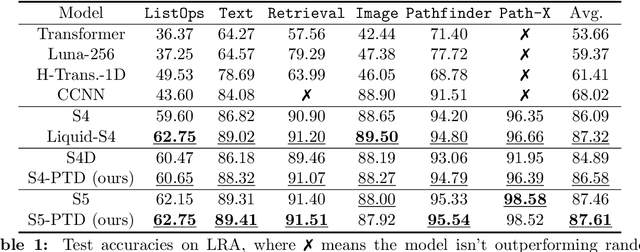
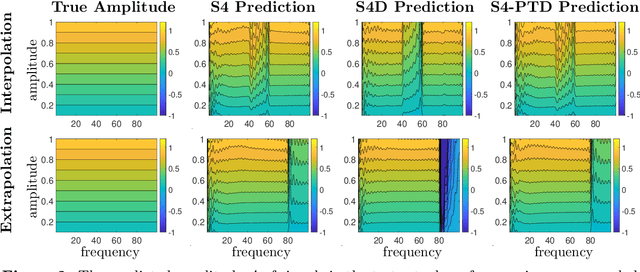
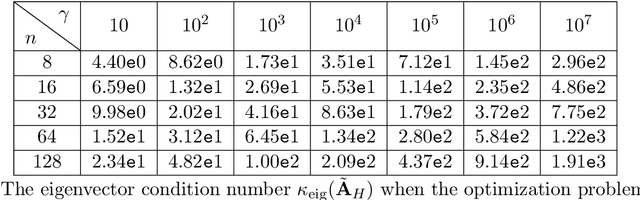
Abstract:State-space models (SSMs) have recently emerged as a framework for learning long-range sequence tasks. An example is the structured state-space sequence (S4) layer, which uses the diagonal-plus-low-rank structure of the HiPPO initialization framework. However, the complicated structure of the S4 layer poses challenges; and, in an effort to address these challenges, models such as S4D and S5 have considered a purely diagonal structure. This choice simplifies the implementation, improves computational efficiency, and allows channel communication. However, diagonalizing the HiPPO framework is itself an ill-posed problem. In this paper, we propose a general solution for this and related ill-posed diagonalization problems in machine learning. We introduce a generic, backward-stable "perturb-then-diagonalize" (PTD) methodology, which is based on the pseudospectral theory of non-normal operators, and which may be interpreted as the approximate diagonalization of the non-normal matrices defining SSMs. Based on this, we introduce the S4-PTD and S5-PTD models. Through theoretical analysis of the transfer functions of different initialization schemes, we demonstrate that the S4-PTD/S5-PTD initialization strongly converges to the HiPPO framework, while the S4D/S5 initialization only achieves weak convergences. As a result, our new models show resilience to Fourier-mode noise-perturbed inputs, a crucial property not achieved by the S4D/S5 models. In addition to improved robustness, our S5-PTD model averages 87.6% accuracy on the Long-Range Arena benchmark, demonstrating that the PTD methodology helps to improve the accuracy of deep learning models.
Towards Foundation Models for Scientific Machine Learning: Characterizing Scaling and Transfer Behavior
Jun 01, 2023



Abstract:Pre-trained machine learning (ML) models have shown great performance for a wide range of applications, in particular in natural language processing (NLP) and computer vision (CV). Here, we study how pre-training could be used for scientific machine learning (SciML) applications, specifically in the context of transfer learning. We study the transfer behavior of these models as (i) the pre-trained model size is scaled, (ii) the downstream training dataset size is scaled, (iii) the physics parameters are systematically pushed out of distribution, and (iv) how a single model pre-trained on a mixture of different physics problems can be adapted to various downstream applications. We find that-when fine-tuned appropriately-transfer learning can help reach desired accuracy levels with orders of magnitude fewer downstream examples (across different tasks that can even be out-of-distribution) than training from scratch, with consistent behavior across a wide range of downstream examples. We also find that fine-tuning these models yields more performance gains as model size increases, compared to training from scratch on new downstream tasks. These results hold for a broad range of PDE learning tasks. All in all, our results demonstrate the potential of the "pre-train and fine-tune" paradigm for SciML problems, demonstrating a path towards building SciML foundation models. We open-source our code for reproducibility.
Topological Regularization via Persistence-Sensitive Optimization
Nov 10, 2020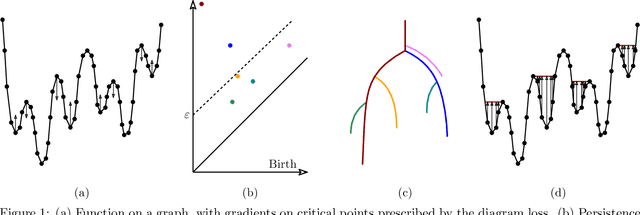


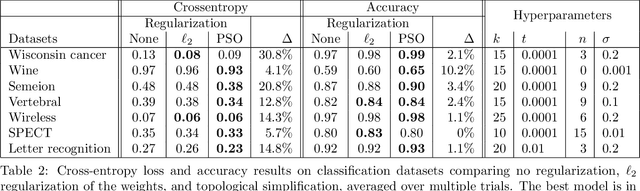
Abstract:Optimization, a key tool in machine learning and statistics, relies on regularization to reduce overfitting. Traditional regularization methods control a norm of the solution to ensure its smoothness. Recently, topological methods have emerged as a way to provide a more precise and expressive control over the solution, relying on persistent homology to quantify and reduce its roughness. All such existing techniques back-propagate gradients through the persistence diagram, which is a summary of the topological features of a function. Their downside is that they provide information only at the critical points of the function. We propose a method that instead builds on persistence-sensitive simplification and translates the required changes to the persistence diagram into changes on large subsets of the domain, including both critical and regular points. This approach enables a faster and more precise topological regularization, the benefits of which we illustrate with experimental evidence.
PersGNN: Applying Topological Data Analysis and Geometric Deep Learning to Structure-Based Protein Function Prediction
Oct 30, 2020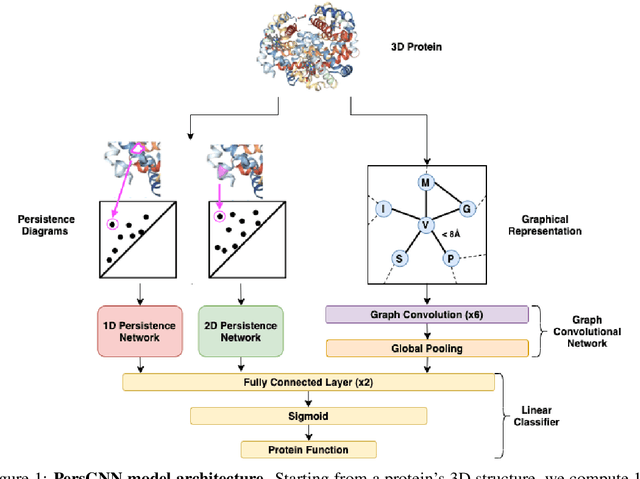
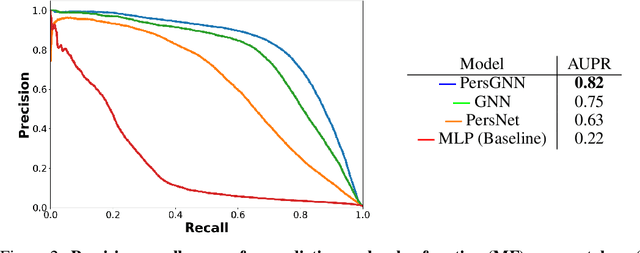
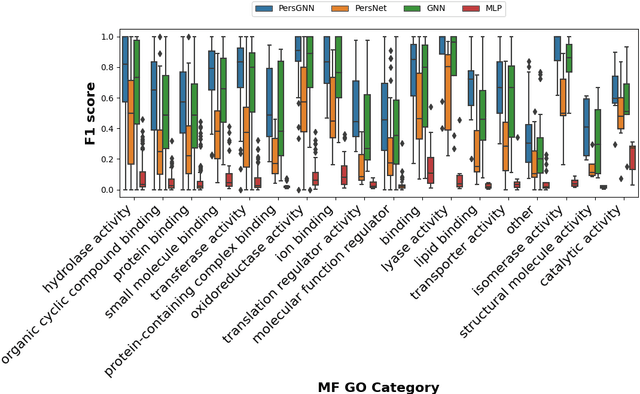
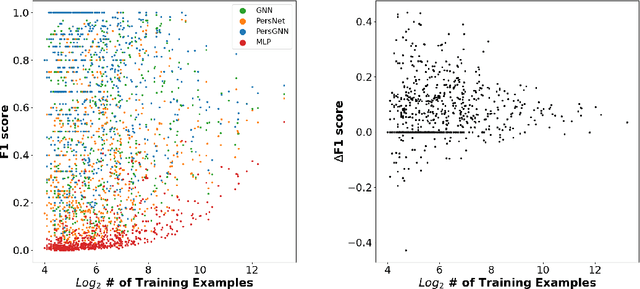
Abstract:Understanding protein structure-function relationships is a key challenge in computational biology, with applications across the biotechnology and pharmaceutical industries. While it is known that protein structure directly impacts protein function, many functional prediction tasks use only protein sequence. In this work, we isolate protein structure to make functional annotations for proteins in the Protein Data Bank in order to study the expressiveness of different structure-based prediction schemes. We present PersGNN - an end-to-end trainable deep learning model that combines graph representation learning with topological data analysis to capture a complex set of both local and global structural features. While variations of these techniques have been successfully applied to proteins before, we demonstrate that our hybridized approach, PersGNN, outperforms either method on its own as well as a baseline neural network that learns from the same information. PersGNN achieves a 9.3% boost in area under the precision recall curve (AUPR) compared to the best individual model, as well as high F1 scores across different gene ontology categories, indicating the transferability of this approach.
Persistent homology advances interpretable machine learning for nanoporous materials
Oct 01, 2020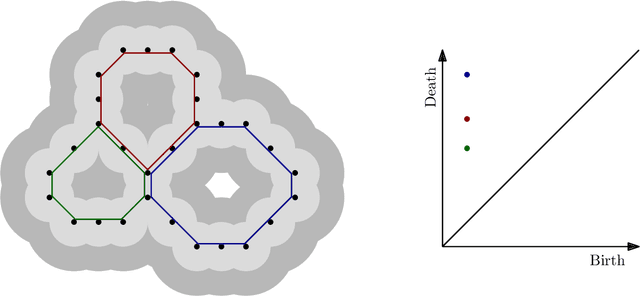
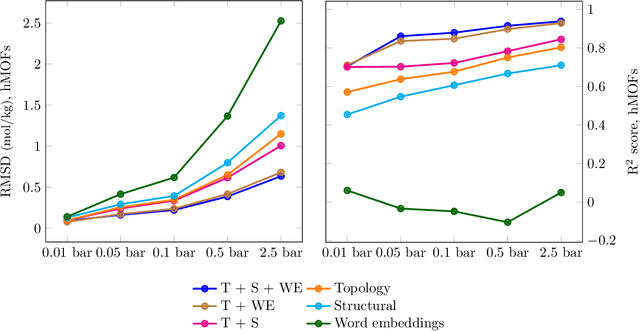

Abstract:Machine learning for nanoporous materials design and discovery has emerged as a promising alternative to more time-consuming experiments and simulations. The challenge with this approach is the selection of features that enable universal and interpretable materials representations across multiple prediction tasks. We use persistent homology to construct holistic representations of the materials structure. We show that these representations can also be augmented with other generic features such as word embeddings from natural language processing to capture chemical information. We demonstrate our approach on multiple metal-organic framework datasets by predicting a variety of gas adsorption targets. Our results show considerable improvement in both accuracy and transferability across targets compared to models constructed from commonly used manually curated features. Persistent homology features allow us to locate the pores that correlate best to adsorption at different pressures, contributing to understanding atomic level structure-property relationships for materials design.
Topological Descriptors Help Predict Guest Adsorption in Nanoporous Materials
Feb 27, 2020

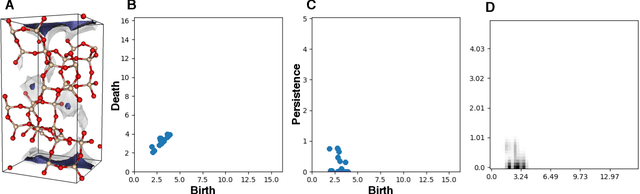
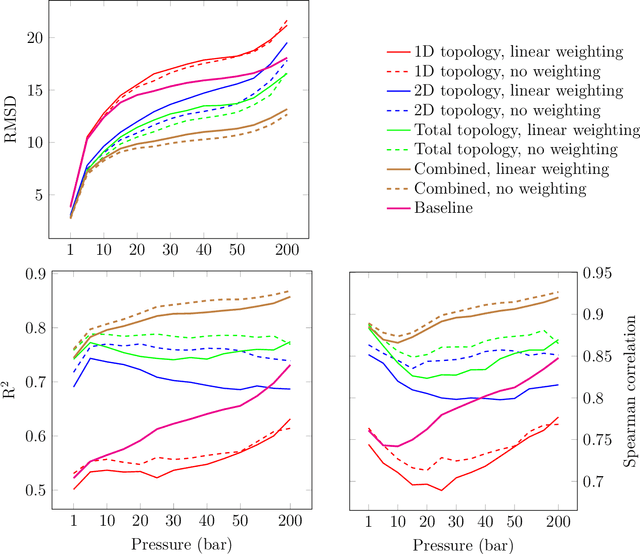
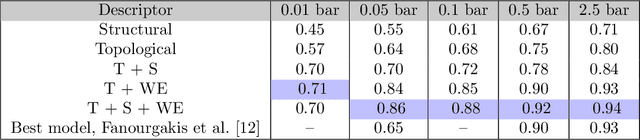
Abstract:Machine learning has emerged as an attractive alternative to experiments and simulations for predicting material properties. Usually, such an approach relies on specific domain knowledge for feature design: each learning target requires careful selection of features that an expert recognizes as important for the specific task. The major drawback of this approach is that computation of only a few structural features has been implemented so far, and it is difficult to tell a priori which features are important for a particular application. The latter problem has been empirically observed for predictors of guest uptake in nanoporous materials: local and global porosity features become dominant descriptors at low and high pressures, respectively. We investigate a feature representation of materials using tools from topological data analysis. Specifically, we use persistent homology to describe the geometry of nanoporous materials at various scales. We combine our topological descriptor with traditional structural features and investigate the relative importance of each to the prediction tasks. We demonstrate an application of this feature representation by predicting methane adsorption in zeolites, for pressures in the range of 1-200 bar. Our results not only show a considerable improvement compared to the baseline, but they also highlight that topological features capture information complementary to the structural features, which is especially important for the adsorption at low pressure, a task particularly difficult for the traditional features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge