Liver Segmentation
Papers and Code
Automating Sonologists USG Commands with AI and Voice Interface
Nov 20, 2024



This research presents an advanced AI-powered ultrasound imaging system that incorporates real-time image processing, organ tracking, and voice commands to enhance the efficiency and accuracy of diagnoses in clinical practice. Traditional ultrasound diagnostics often require significant time and introduce a degree of subjectivity due to user interaction. The goal of this innovative solution is to provide Sonologists with a more predictable and productive imaging procedure utilizing artificial intelligence, computer vision, and voice technology. The functionality of the system employs computer vision and deep learning algorithms, specifically adopting the Mask R-CNN model from Detectron2 for semantic segmentation of organs and key landmarks. This automation improves diagnostic accuracy by enabling the extraction of valuable information with minimal human input. Additionally, it includes a voice recognition feature that allows for hands-free operation, enabling users to control the system with commands such as freeze or liver, all while maintaining their focus on the patient. The architecture comprises video processing and real-time segmentation modules that prepare the system to perform essential imaging functions, such as freezing and zooming in on frames. The liver histopathology module, optimized for detecting fibrosis, achieved an impressive accuracy of 98.6%. Furthermore, the organ segmentation module produces output confidence levels between 50% and 95%, demonstrating its efficacy in organ detection.
MiniGPT-Pancreas: Multimodal Large Language Model for Pancreas Cancer Classification and Detection
Dec 20, 2024

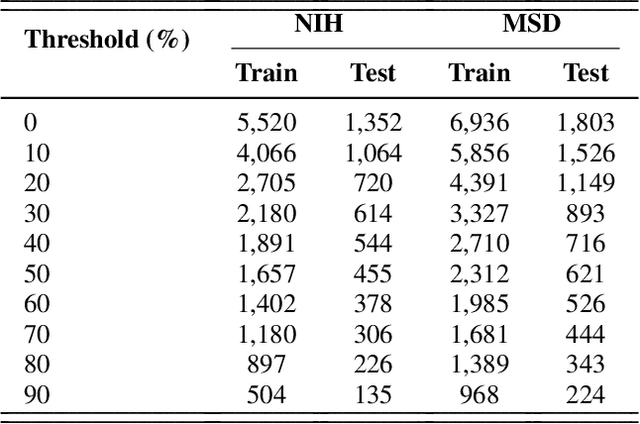
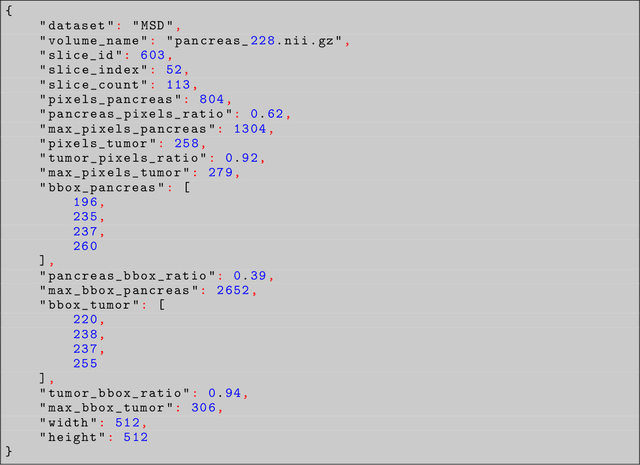
Problem: Pancreas radiological imaging is challenging due to the small size, blurred boundaries, and variability of shape and position of the organ among patients. Goal: In this work we present MiniGPT-Pancreas, a Multimodal Large Language Model (MLLM), as an interactive chatbot to support clinicians in pancreas cancer diagnosis by integrating visual and textual information. Methods: MiniGPT-v2, a general-purpose MLLM, was fine-tuned in a cascaded way for pancreas detection, tumor classification, and tumor detection with multimodal prompts combining questions and computed tomography scans from the National Institute of Health (NIH), and Medical Segmentation Decathlon (MSD) datasets. The AbdomenCT-1k dataset was used to detect the liver, spleen, kidney, and pancreas. Results: MiniGPT-Pancreas achieved an Intersection over Union (IoU) of 0.595 and 0.550 for the detection of pancreas on NIH and MSD datasets, respectively. For the pancreas cancer classification task on the MSD dataset, accuracy, precision, and recall were 0.876, 0.874, and 0.878, respectively. When evaluating MiniGPT-Pancreas on the AbdomenCT-1k dataset for multi-organ detection, the IoU was 0.8399 for the liver, 0.722 for the kidney, 0.705 for the spleen, and 0.497 for the pancreas. For the pancreas tumor detection task, the IoU score was 0.168 on the MSD dataset. Conclusions: MiniGPT-Pancreas represents a promising solution to support clinicians in the classification of pancreas images with pancreas tumors. Future research is needed to improve the score on the detection task, especially for pancreas tumors.
Towards Synergistic Deep Learning Models for Volumetric Cirrhotic Liver Segmentation in MRIs
Aug 08, 2024



Liver cirrhosis, a leading cause of global mortality, requires precise segmentation of ROIs for effective disease monitoring and treatment planning. Existing segmentation models often fail to capture complex feature interactions and generalize across diverse datasets. To address these limitations, we propose a novel synergistic theory that leverages complementary latent spaces for enhanced feature interaction modeling. Our proposed architecture, nnSynergyNet3D integrates continuous and discrete latent spaces for 3D volumes and features auto-configured training. This approach captures both fine-grained and coarse features, enabling effective modeling of intricate feature interactions. We empirically validated nnSynergyNet3D on a private dataset of 628 high-resolution T1 abdominal MRI scans from 339 patients. Our model outperformed the baseline nnUNet3D by approximately 2%. Additionally, zero-shot testing on healthy liver CT scans from the public LiTS dataset demonstrated superior cross-modal generalization capabilities. These results highlight the potential of synergistic latent space models to improve segmentation accuracy and robustness, thereby enhancing clinical workflows by ensuring consistency across CT and MRI modalities.
Spectral U-Net: Enhancing Medical Image Segmentation via Spectral Decomposition
Sep 13, 2024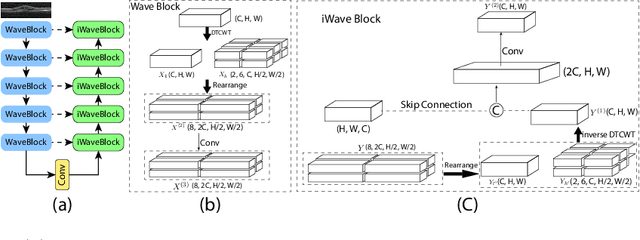

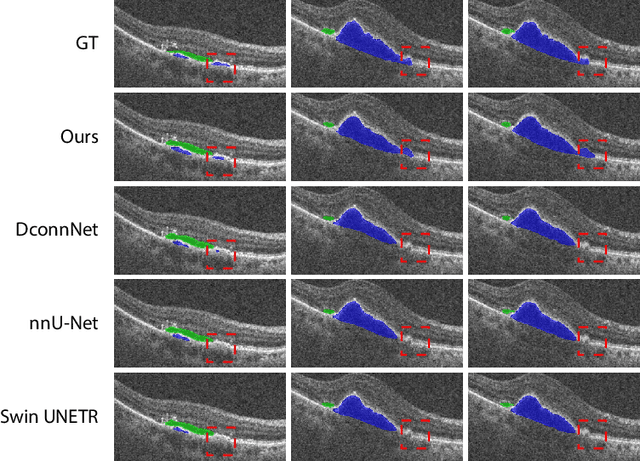

This paper introduces Spectral U-Net, a novel deep learning network based on spectral decomposition, by exploiting Dual Tree Complex Wavelet Transform (DTCWT) for down-sampling and inverse Dual Tree Complex Wavelet Transform (iDTCWT) for up-sampling. We devise the corresponding Wave-Block and iWave-Block, integrated into the U-Net architecture, aiming at mitigating information loss during down-sampling and enhancing detail reconstruction during up-sampling. In the encoder, we first decompose the feature map into high and low-frequency components using DTCWT, enabling down-sampling while mitigating information loss. In the decoder, we utilize iDTCWT to reconstruct higher-resolution feature maps from down-sampled features. Evaluations on the Retina Fluid, Brain Tumor, and Liver Tumor segmentation datasets with the nnU-Net framework demonstrate the superiority of the proposed Spectral U-Net.
A Novel Automatic Real-time Motion Tracking Method for Magnetic Resonance Imaging-guided Radiotherapy: Leveraging the Enhanced Tracking-Learning-Detection Framework with Automatic Segmentation
Nov 12, 2024



Objective: Ensuring the precision in motion tracking for MRI-guided Radiotherapy (MRIgRT) is crucial for the delivery of effective treatments. This study refined the motion tracking accuracy in MRIgRT through the innovation of an automatic real-time tracking method, leveraging an enhanced Tracking-Learning-Detection (ETLD) framework coupled with automatic segmentation. Methods: We developed a novel MRIgRT motion tracking method by integrating two primary methods: the ETLD framework and an improved Chan-Vese model (ICV), named ETLD+ICV. The TLD framework was upgraded to suit real-time cine MRI, including advanced image preprocessing, no-reference image quality assessment, an enhanced median-flow tracker, and a refined detector with dynamic search region adjustments. Additionally, ICV was combined for precise coverage of the target volume, which refined the segmented region frame by frame using tracking results, with key parameters optimized. Tested on 3.5D MRI scans from 10 patients with liver metastases, our method ensures precise tracking and accurate segmentation vital for MRIgRT. Results: An evaluation of 106,000 frames across 77 treatment fractions revealed sub-millimeter tracking errors of less than 0.8mm, with over 99% precision and 98% recall for all subjects, underscoring the robustness and efficacy of the ETLD. Moreover, the ETLD+ICV yielded a dice global score of more than 82% for all subjects, demonstrating the proposed method's extensibility and precise target volume coverage. Conclusions: This study successfully developed an automatic real-time motion tracking method for MRIgRT that markedly surpasses current methods. The novel method not only delivers exceptional precision in tracking and segmentation but also demonstrates enhanced adaptability to clinical demands, positioning it as an indispensable asset in the quest to augment the efficacy of radiotherapy treatments.
Expanded Comprehensive Robotic Cholecystectomy Dataset (CRCD)
Dec 16, 2024



In recent years, the application of machine learning to minimally invasive surgery (MIS) has attracted considerable interest. Datasets are critical to the use of such techniques. This paper presents a unique dataset recorded during ex vivo pseudo-cholecystectomy procedures on pig livers using the da Vinci Research Kit (dVRK). Unlike existing datasets, it addresses a critical gap by providing comprehensive kinematic data, recordings of all pedal inputs, and offers a time-stamped record of the endoscope's movements. This expanded version also includes segmentation and keypoint annotations of images, enhancing its utility for computer vision applications. Contributed by seven surgeons with varied backgrounds and experience levels that are provided as a part of this expanded version, the dataset is an important new resource for surgical robotics research. It enables the development of advanced methods for evaluating surgeon skills, tools for providing better context awareness, and automation of surgical tasks. Our work overcomes the limitations of incomplete recordings and imprecise kinematic data found in other datasets. To demonstrate the potential of the dataset for advancing automation in surgical robotics, we introduce two models that predict clutch usage and camera activation, a 3D scene reconstruction example, and the results from our keypoint and segmentation models.
A Novel Momentum-Based Deep Learning Techniques for Medical Image Classification and Segmentation
Aug 11, 2024



Accurately segmenting different organs from medical images is a critical prerequisite for computer-assisted diagnosis and intervention planning. This study proposes a deep learning-based approach for segmenting various organs from CT and MRI scans and classifying diseases. Our study introduces a novel technique integrating momentum within residual blocks for enhanced training dynamics in medical image analysis. We applied our method in two distinct tasks: segmenting liver, lung, & colon data and classifying abdominal pelvic CT and MRI scans. The proposed approach has shown promising results, outperforming state-of-the-art methods on publicly available benchmarking datasets. For instance, in the lung segmentation dataset, our approach yielded significant enhancements over the TransNetR model, including a 5.72% increase in dice score, a 5.04% improvement in mean Intersection over Union (mIoU), an 8.02% improvement in recall, and a 4.42% improvement in precision. Hence, incorporating momentum led to state-of-the-art performance in both segmentation and classification tasks, representing a significant advancement in the field of medical imaging.
SMAFormer: Synergistic Multi-Attention Transformer for Medical Image Segmentation
Aug 31, 2024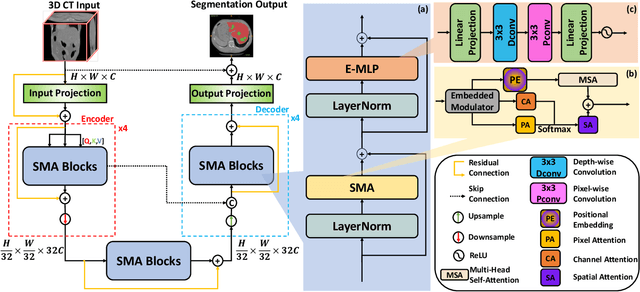
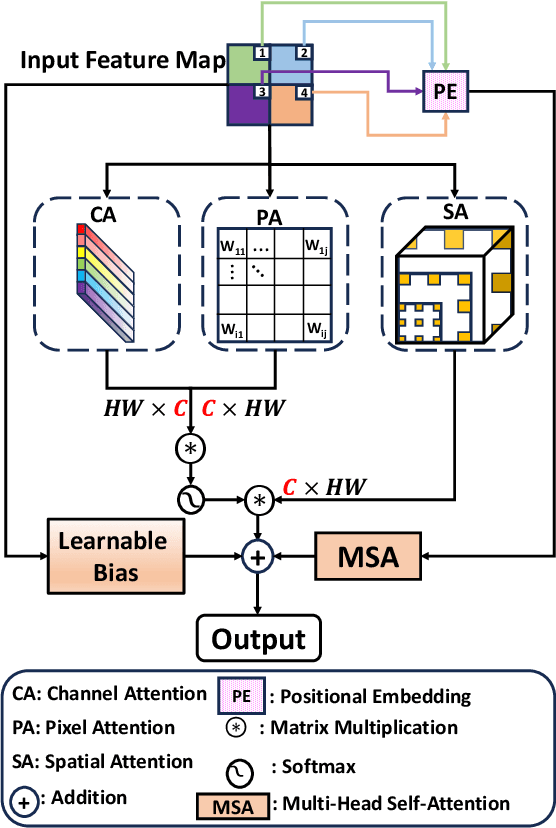
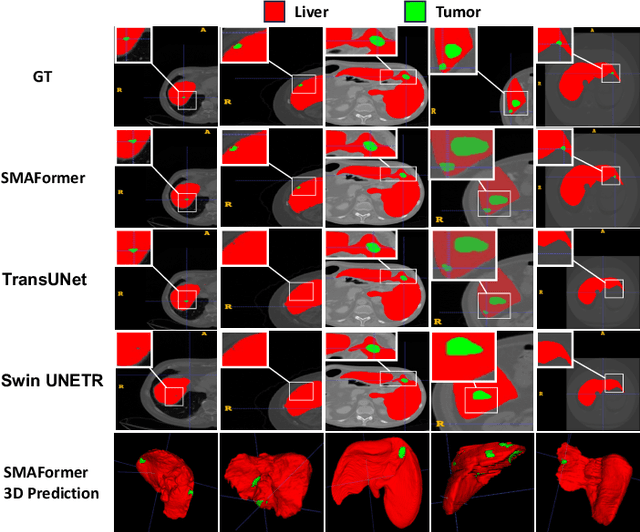
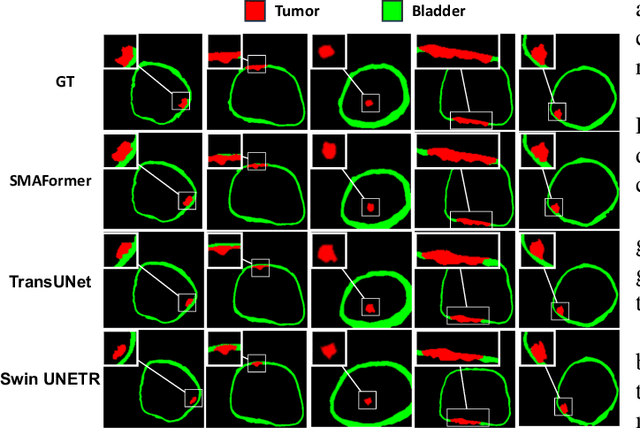
In medical image segmentation, specialized computer vision techniques, notably transformers grounded in attention mechanisms and residual networks employing skip connections, have been instrumental in advancing performance. Nonetheless, previous models often falter when segmenting small, irregularly shaped tumors. To this end, we introduce SMAFormer, an efficient, Transformer-based architecture that fuses multiple attention mechanisms for enhanced segmentation of small tumors and organs. SMAFormer can capture both local and global features for medical image segmentation. The architecture comprises two pivotal components. First, a Synergistic Multi-Attention (SMA) Transformer block is proposed, which has the benefits of Pixel Attention, Channel Attention, and Spatial Attention for feature enrichment. Second, addressing the challenge of information loss incurred during attention mechanism transitions and feature fusion, we design a Feature Fusion Modulator. This module bolsters the integration between the channel and spatial attention by mitigating reshaping-induced information attrition. To evaluate our method, we conduct extensive experiments on various medical image segmentation tasks, including multi-organ, liver tumor, and bladder tumor segmentation, achieving state-of-the-art results. Code and models are available at: \url{https://github.com/CXH-Research/SMAFormer}.
ASSNet: Adaptive Semantic Segmentation Network for Microtumors and Multi-Organ Segmentation
Sep 12, 2024



Medical image segmentation, a crucial task in computer vision, facilitates the automated delineation of anatomical structures and pathologies, supporting clinicians in diagnosis, treatment planning, and disease monitoring. Notably, transformers employing shifted window-based self-attention have demonstrated exceptional performance. However, their reliance on local window attention limits the fusion of local and global contextual information, crucial for segmenting microtumors and miniature organs. To address this limitation, we propose the Adaptive Semantic Segmentation Network (ASSNet), a transformer architecture that effectively integrates local and global features for precise medical image segmentation. ASSNet comprises a transformer-based U-shaped encoder-decoder network. The encoder utilizes shifted window self-attention across five resolutions to extract multi-scale features, which are then propagated to the decoder through skip connections. We introduce an augmented multi-layer perceptron within the encoder to explicitly model long-range dependencies during feature extraction. Recognizing the constraints of conventional symmetrical encoder-decoder designs, we propose an Adaptive Feature Fusion (AFF) decoder to complement our encoder. This decoder incorporates three key components: the Long Range Dependencies (LRD) block, the Multi-Scale Feature Fusion (MFF) block, and the Adaptive Semantic Center (ASC) block. These components synergistically facilitate the effective fusion of multi-scale features extracted by the decoder while capturing long-range dependencies and refining object boundaries. Comprehensive experiments on diverse medical image segmentation tasks, including multi-organ, liver tumor, and bladder tumor segmentation, demonstrate that ASSNet achieves state-of-the-art results. Code and models are available at: \url{https://github.com/lzeeorno/ASSNet}.
AI Radiologist: Revolutionizing Liver Tissue Segmentation with Convolutional Neural Networks and a Clinician-Friendly GUI
Jun 11, 2024



Artificial Intelligence (AI) is a pervasive research topic, permeating various sectors and applications. In this study, we harness the power of AI, specifically convolutional neural networks (ConvNets), for segmenting liver tissues. It also focuses on developing a user-friendly graphical user interface (GUI) tool, "AI Radiologist", enabling clinicians to effectively delineate different liver tissues (parenchyma, tumors, and vessels), thereby saving lives. This endeavor bridges the gap between academic research and practical, industrial applications. The GUI is a single-page application and is designed using the PyQt5 Python framework. The offline-available AI Radiologist resorts to three ConvNet models trained to segment all liver tissues. With respect to the Dice metric, the best liver ConvNet scores 98.16%, the best tumor ConvNet scores 65.95%, and the best vessel ConvNet scores 51.94%. It outputs 2D slices of the liver, tumors, and vessels, along with 3D interpolations in .obj and .mtl formats, which can be visualized/printed using any 3D-compatible software. Thus, the AI Radiologist offers a convenient tool for clinicians to perform liver tissue segmentation and 3D interpolation employing state-of-the-art models for tissues segmentation. With the provided capacity to select the volumes and pre-trained models, the clinicians can leave the rest to the AI Radiologist.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge