Shoujun Zhou
DV-VLN: Dual Verification for Reliable LLM-Based Vision-and-Language Navigation
Jan 26, 2026Abstract:Vision-and-Language Navigation (VLN) requires an embodied agent to navigate in a complex 3D environment according to natural language instructions. Recent progress in large language models (LLMs) has enabled language-driven navigation with improved interpretability. However, most LLM-based agents still rely on single-shot action decisions, where the model must choose one option from noisy, textualized multi-perspective observations. Due to local mismatches and imperfect intermediate reasoning, such decisions can easily deviate from the correct path, leading to error accumulation and reduced reliability in unseen environments. In this paper, we propose DV-VLN, a new VLN framework that follows a generate-then-verify paradigm. DV-VLN first performs parameter-efficient in-domain adaptation of an open-source LLaMA-2 backbone to produce a structured navigational chain-of-thought, and then verifies candidate actions with two complementary channels: True-False Verification (TFV) and Masked-Entity Verification (MEV). DV-VLN selects actions by aggregating verification successes across multiple samples, yielding interpretable scores for reranking. Experiments on R2R, RxR (English subset), and REVERIE show that DV-VLN consistently improves over direct prediction and sampling-only baselines, achieving competitive performance among language-only VLN agents and promising results compared with several cross-modal systems.Code is available at https://github.com/PlumJun/DV-VLN.
Adaptive Morph-Patch Transformer for Aortic Vessel Segmentation
Nov 11, 2025Abstract:Accurate segmentation of aortic vascular structures is critical for diagnosing and treating cardiovascular diseases.Traditional Transformer-based models have shown promise in this domain by capturing long-range dependencies between vascular features. However, their reliance on fixed-size rectangular patches often influences the integrity of complex vascular structures, leading to suboptimal segmentation accuracy. To address this challenge, we propose the adaptive Morph Patch Transformer (MPT), a novel architecture specifically designed for aortic vascular segmentation. Specifically, MPT introduces an adaptive patch partitioning strategy that dynamically generates morphology-aware patches aligned with complex vascular structures. This strategy can preserve semantic integrity of complex vascular structures within individual patches. Moreover, a Semantic Clustering Attention (SCA) method is proposed to dynamically aggregate features from various patches with similar semantic characteristics. This method enhances the model's capability to segment vessels of varying sizes, preserving the integrity of vascular structures. Extensive experiments on three open-source dataset(AVT, AortaSeg24 and TBAD) demonstrate that MPT achieves state-of-the-art performance, with improvements in segmenting intricate vascular structures.
Overview of the NLPCC 2025 Shared Task 4: Multi-modal, Multilingual, and Multi-hop Medical Instructional Video Question Answering Challenge
May 11, 2025Abstract:Following the successful hosts of the 1-st (NLPCC 2023 Foshan) CMIVQA and the 2-rd (NLPCC 2024 Hangzhou) MMIVQA challenges, this year, a new task has been introduced to further advance research in multi-modal, multilingual, and multi-hop medical instructional question answering (M4IVQA) systems, with a specific focus on medical instructional videos. The M4IVQA challenge focuses on evaluating models that integrate information from medical instructional videos, understand multiple languages, and answer multi-hop questions requiring reasoning over various modalities. This task consists of three tracks: multi-modal, multilingual, and multi-hop Temporal Answer Grounding in Single Video (M4TAGSV), multi-modal, multilingual, and multi-hop Video Corpus Retrieval (M4VCR) and multi-modal, multilingual, and multi-hop Temporal Answer Grounding in Video Corpus (M4TAGVC). Participants in M4IVQA are expected to develop algorithms capable of processing both video and text data, understanding multilingual queries, and providing relevant answers to multi-hop medical questions. We believe the newly introduced M4IVQA challenge will drive innovations in multimodal reasoning systems for healthcare scenarios, ultimately contributing to smarter emergency response systems and more effective medical education platforms in multilingual communities. Our official website is https://cmivqa.github.io/
ReGraP-LLaVA: Reasoning enabled Graph-based Personalized Large Language and Vision Assistant
May 06, 2025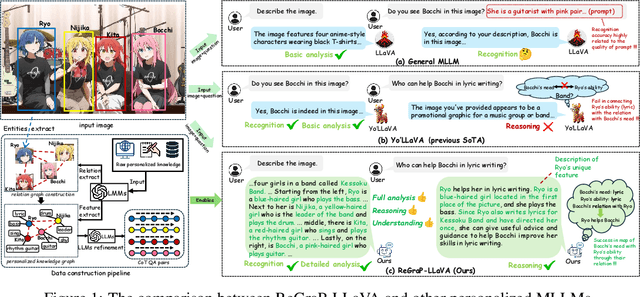

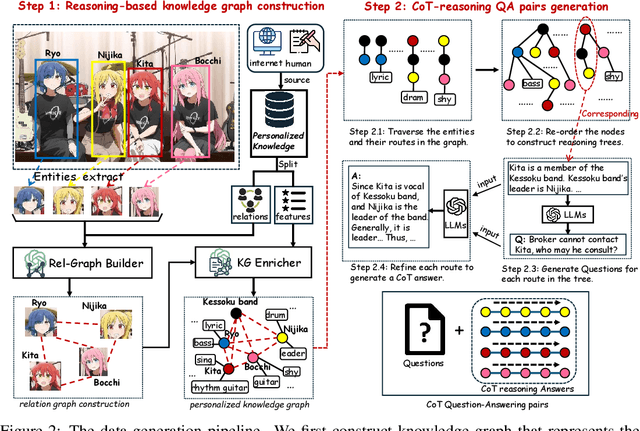
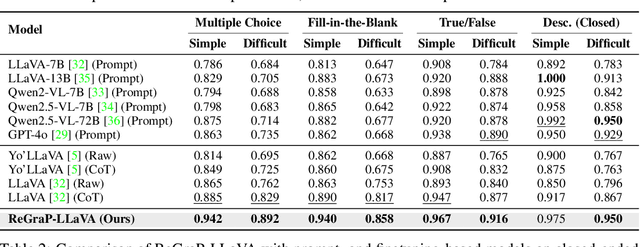
Abstract:Recent advances in personalized MLLMs enable effective capture of user-specific concepts, supporting both recognition of personalized concepts and contextual captioning. However, humans typically explore and reason over relations among objects and individuals, transcending surface-level information to achieve more personalized and contextual understanding. To this end, existing methods may face three main limitations: Their training data lacks multi-object sets in which relations among objects are learnable. Building on the limited training data, their models overlook the relations between different personalized concepts and fail to reason over them. Their experiments mainly focus on a single personalized concept, where evaluations are limited to recognition and captioning tasks. To address the limitations, we present a new dataset named ReGraP, consisting of 120 sets of personalized knowledge. Each set includes images, KGs, and CoT QA pairs derived from the KGs, enabling more structured and sophisticated reasoning pathways. We propose ReGraP-LLaVA, an MLLM trained with the corresponding KGs and CoT QA pairs, where soft and hard graph prompting methods are designed to align KGs within the model's semantic space. We establish the ReGraP Benchmark, which contains diverse task types: multiple-choice, fill-in-the-blank, True/False, and descriptive questions in both open- and closed-ended settings. The proposed benchmark is designed to evaluate the relational reasoning and knowledge-connection capability of personalized MLLMs. We conduct experiments on the proposed ReGraP-LLaVA and other competitive MLLMs. Results show that the proposed model not only learns personalized knowledge but also performs relational reasoning in responses, achieving the SoTA performance compared with the competitive methods. All the codes and datasets are released at: https://github.com/xyfyyds/ReGraP.
Ask2Loc: Learning to Locate Instructional Visual Answers by Asking Questions
Apr 22, 2025Abstract:Locating specific segments within an instructional video is an efficient way to acquire guiding knowledge. Generally, the task of obtaining video segments for both verbal explanations and visual demonstrations is known as visual answer localization (VAL). However, users often need multiple interactions to obtain answers that align with their expectations when using the system. During these interactions, humans deepen their understanding of the video content by asking themselves questions, thereby accurately identifying the location. Therefore, we propose a new task, named In-VAL, to simulate the multiple interactions between humans and videos in the procedure of obtaining visual answers. The In-VAL task requires interactively addressing several semantic gap issues, including 1) the ambiguity of user intent in the input questions, 2) the incompleteness of language in video subtitles, and 3) the fragmentation of content in video segments. To address these issues, we propose Ask2Loc, a framework for resolving In-VAL by asking questions. It includes three key modules: 1) a chatting module to refine initial questions and uncover clear intentions, 2) a rewriting module to generate fluent language and create complete descriptions, and 3) a searching module to broaden local context and provide integrated content. We conduct extensive experiments on three reconstructed In-VAL datasets. Compared to traditional end-to-end and two-stage methods, our proposed Ask2Loc can improve performance by up to 14.91 (mIoU) on the In-VAL task. Our code and datasets can be accessed at https://github.com/changzong/Ask2Loc.
Hierarchical Modeling for Medical Visual Question Answering with Cross-Attention Fusion
Apr 10, 2025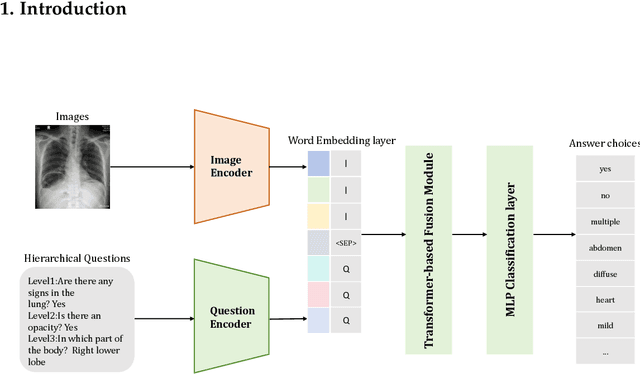
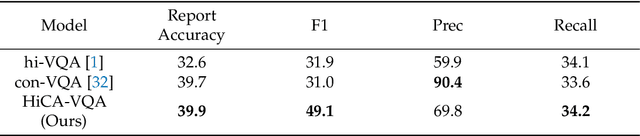
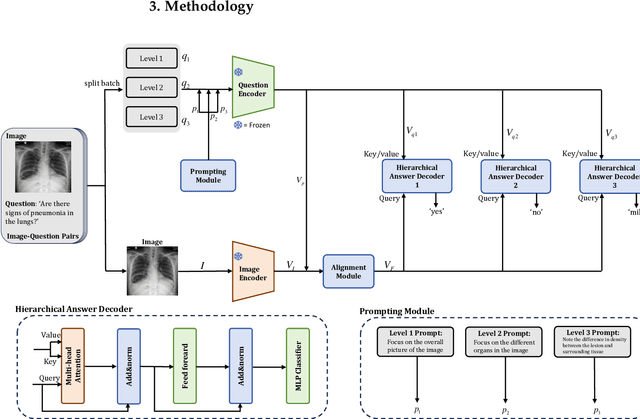
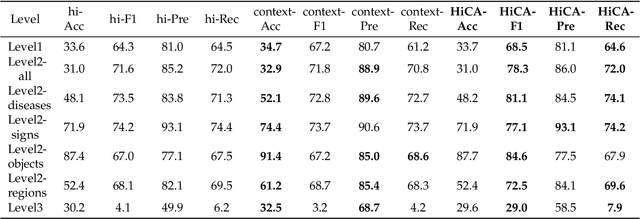
Abstract:Medical Visual Question Answering (Med-VQA) answers clinical questions using medical images, aiding diagnosis. Designing the MedVQA system holds profound importance in assisting clinical diagnosis and enhancing diagnostic accuracy. Building upon this foundation, Hierarchical Medical VQA extends Medical VQA by organizing medical questions into a hierarchical structure and making level-specific predictions to handle fine-grained distinctions. Recently, many studies have proposed hierarchical MedVQA tasks and established datasets, However, several issues still remain: (1) imperfect hierarchical modeling leads to poor differentiation between question levels causing semantic fragmentation across hierarchies. (2) Excessive reliance on implicit learning in Transformer-based cross-modal self-attention fusion methods, which obscures crucial local semantic correlations in medical scenarios. To address these issues, this study proposes a HiCA-VQA method, including two modules: Hierarchical Prompting for fine-grained medical questions and Hierarchical Answer Decoders. The hierarchical prompting module pre-aligns hierarchical text prompts with image features to guide the model in focusing on specific image regions according to question types, while the hierarchical decoder performs separate predictions for questions at different levels to improve accuracy across granularities. The framework also incorporates a cross-attention fusion module where images serve as queries and text as key-value pairs. Experiments on the Rad-Restruct benchmark demonstrate that the HiCA-VQA framework better outperforms existing state-of-the-art methods in answering hierarchical fine-grained questions. This study provides an effective pathway for hierarchical visual question answering systems, advancing medical image understanding.
MSA-UNet3+: Multi-Scale Attention UNet3+ with New Supervised Prototypical Contrastive Loss for Coronary DSA Image Segmentation
Apr 07, 2025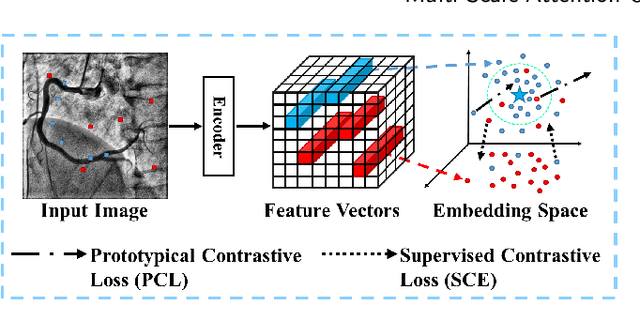

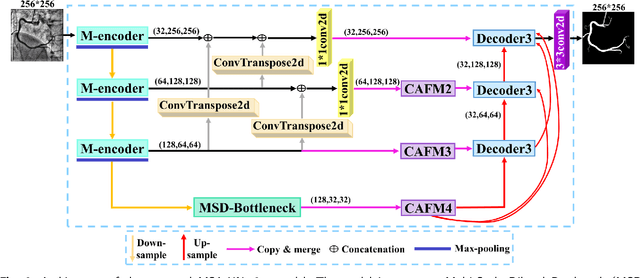
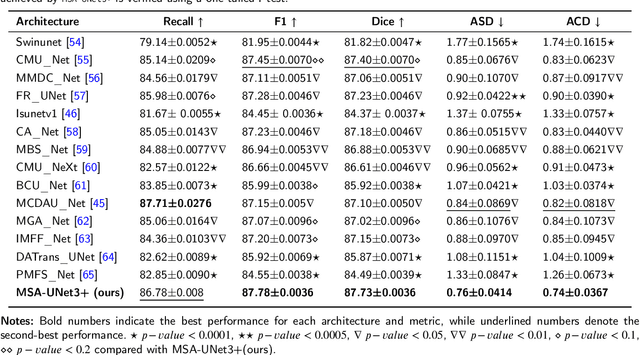
Abstract:The accurate segmentation of coronary Digital Subtraction Angiography (DSA) images is essential for diagnosing and treating coronary artery diseases. Despite advances in deep learning-based segmentation, challenges such as low contrast, noise, overlapping structures, high intra-class variance, and class imbalance limit precise vessel delineation. To overcome these limitations, we propose the MSA-UNet3+: a Multi-Scale Attention enhanced UNet3+ architecture for coronary DSA image segmentation. The framework combined Multi-Scale Dilated Bottleneck (MSD-Bottleneck) with Contextual Attention Fusion Module (CAFM), which not only enhances multi-scale feature extraction but also preserve fine-grained details, and improve contextual understanding. Furthermore, we propose a new Supervised Prototypical Contrastive Loss (SPCL), which combines supervised and prototypical contrastive learning to minimize class imbalance and high intra-class variance by focusing on hard-to-classified background samples. Experiments carried out on a private coronary DSA dataset demonstrate that MSA-UNet3+ outperforms state-of-the-art methods, achieving a Dice coefficient of 87.73%, an F1-score of 87.78%, and significantly reduced Average Surface Distance (ASD) and Average Contour Distance (ACD). The developed framework provides clinicians with precise vessel segmentation, enabling accurate identification of coronary stenosis and supporting informed diagnostic and therapeutic decisions. The code will be released at the following GitHub profile link https://github.com/rayanmerghani/MSA-UNet3plus.
Small but Mighty: Enhancing Time Series Forecasting with Lightweight LLMs
Mar 05, 2025Abstract:While LLMs have demonstrated remarkable potential in time series forecasting, their practical deployment remains constrained by excessive computational demands and memory footprints. Existing LLM-based approaches typically suffer from three critical limitations: Inefficient parameter utilization in handling numerical time series patterns; Modality misalignment between continuous temporal signals and discrete text embeddings; and Inflexibility for real-time expert knowledge integration. We present SMETimes, the first systematic investigation of sub-3B parameter SLMs for efficient and accurate time series forecasting. Our approach centers on three key innovations: A statistically-enhanced prompting mechanism that bridges numerical time series with textual semantics through descriptive statistical features; A adaptive fusion embedding architecture that aligns temporal patterns with language model token spaces through learnable parameters; And a dynamic mixture-of-experts framework enabled by SLMs' computational efficiency, adaptively combining base predictions with domain-specific models. Extensive evaluations across seven benchmark datasets demonstrate that our 3B-parameter SLM achieves state-of-the-art performance on five primary datasets while maintaining 3.8x faster training and 5.2x lower memory consumption compared to 7B-parameter LLM baselines. Notably, the proposed model exhibits better learning capabilities, achieving 12.3% lower MSE than conventional LLM. Ablation studies validate that our statistical prompting and cross-modal fusion modules respectively contribute 15.7% and 18.2% error reduction in long-horizon forecasting tasks. By redefining the efficiency-accuracy trade-off landscape, this work establishes SLMs as viable alternatives to resource-intensive LLMs for practical time series forecasting. Code and models are available at https://github.com/xiyan1234567/SMETimes.
VesselSAM: Leveraging SAM for Aortic Vessel Segmentation with LoRA and Atrous Attention
Feb 25, 2025Abstract:Medical image segmentation is crucial for clinical diagnosis and treatment planning, particularly for complex anatomical structures like vessels. In this work, we propose VesselSAM, a modified version of the Segmentation Anything Model (SAM), specifically designed for aortic vessel segmentation. VesselSAM incorporates AtrousLoRA, a novel module that combines Atrous Attention with Low-Rank Adaptation (LoRA), to improve segmentation performance. Atrous Attention enables the model to capture multi-scale contextual information, preserving both fine local details and broader global context. At the same time, LoRA facilitates efficient fine-tuning of the frozen SAM image encoder, reducing the number of trainable parameters and ensuring computational efficiency. We evaluate VesselSAM on two challenging datasets: the Aortic Vessel Tree (AVT) dataset and the Type-B Aortic Dissection (TBAD) dataset. VesselSAM achieves state-of-the-art performance with DSC scores of 93.50\%, 93.25\%, 93.02\%, and 93.26\% across multiple medical centers. Our results demonstrate that VesselSAM delivers high segmentation accuracy while significantly reducing computational overhead compared to existing large-scale models. This development paves the way for enhanced AI-based aortic vessel segmentation in clinical environments. The code and models will be released at https://github.com/Adnan-CAS/AtrousLora.
ClinKD: Cross-Modal Clinic Knowledge Distiller For Multi-Task Medical Images
Feb 09, 2025



Abstract:Med-VQA (Medical Visual Question Answering) is a crucial subtask within the broader VQA (Visual Question Answering) domain. This task requires a visual question answering system to analyze the provided image and corresponding question,offering reasonable analysis and suggestions to assist medical professionals in making pathological diagnoses, or ideally, enabling the system to independently provide correct diagnoses. Furthermore, more advanced Med-VQA tasks involve Referring and Grounding, which not only require the system to accurately comprehend medical images but also to pinpoint specific biological locations within those images. While many large pre-trained models have demonstrated substantial VQA capabilities,challenges persist in the medical imaging domain. The intricacy of biological features in medical images and the scarcity of high-quality medical image datasets, combined with the fact that current models are not tailored for the medical field in terms of architecture and training paradigms, hinder the full exploitation of model generalization. This results in issues such as hallucination in Visual Grounding. In this paper, we introduce the ClinKD model, which incorporates modifications to model position encoding and a diversified training process. Initially, we enhance the model's ability to perceive image and modality variations by using Med-CLIP Guided Rotary Position Embedding. Subsequently, we leverage distillation to provide prior knowledge to the model before using complete training data. Additionally, the feedback-based training process during the formal training phase further enhances data utilization. Notably, under unchanged evaluation protocols, we achieve a new state-of-the-art performance on the Med-GRIT-270k dataset, and the Med-CLIP Guided Rotary Position Embedding approach presents potential for generalizing to universal model position encoding.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge