Zhenxi Zhang
DV-VLN: Dual Verification for Reliable LLM-Based Vision-and-Language Navigation
Jan 26, 2026Abstract:Vision-and-Language Navigation (VLN) requires an embodied agent to navigate in a complex 3D environment according to natural language instructions. Recent progress in large language models (LLMs) has enabled language-driven navigation with improved interpretability. However, most LLM-based agents still rely on single-shot action decisions, where the model must choose one option from noisy, textualized multi-perspective observations. Due to local mismatches and imperfect intermediate reasoning, such decisions can easily deviate from the correct path, leading to error accumulation and reduced reliability in unseen environments. In this paper, we propose DV-VLN, a new VLN framework that follows a generate-then-verify paradigm. DV-VLN first performs parameter-efficient in-domain adaptation of an open-source LLaMA-2 backbone to produce a structured navigational chain-of-thought, and then verifies candidate actions with two complementary channels: True-False Verification (TFV) and Masked-Entity Verification (MEV). DV-VLN selects actions by aggregating verification successes across multiple samples, yielding interpretable scores for reranking. Experiments on R2R, RxR (English subset), and REVERIE show that DV-VLN consistently improves over direct prediction and sampling-only baselines, achieving competitive performance among language-only VLN agents and promising results compared with several cross-modal systems.Code is available at https://github.com/PlumJun/DV-VLN.
Adaptive Morph-Patch Transformer for Aortic Vessel Segmentation
Nov 11, 2025Abstract:Accurate segmentation of aortic vascular structures is critical for diagnosing and treating cardiovascular diseases.Traditional Transformer-based models have shown promise in this domain by capturing long-range dependencies between vascular features. However, their reliance on fixed-size rectangular patches often influences the integrity of complex vascular structures, leading to suboptimal segmentation accuracy. To address this challenge, we propose the adaptive Morph Patch Transformer (MPT), a novel architecture specifically designed for aortic vascular segmentation. Specifically, MPT introduces an adaptive patch partitioning strategy that dynamically generates morphology-aware patches aligned with complex vascular structures. This strategy can preserve semantic integrity of complex vascular structures within individual patches. Moreover, a Semantic Clustering Attention (SCA) method is proposed to dynamically aggregate features from various patches with similar semantic characteristics. This method enhances the model's capability to segment vessels of varying sizes, preserving the integrity of vascular structures. Extensive experiments on three open-source dataset(AVT, AortaSeg24 and TBAD) demonstrate that MPT achieves state-of-the-art performance, with improvements in segmenting intricate vascular structures.
ReGraP-LLaVA: Reasoning enabled Graph-based Personalized Large Language and Vision Assistant
May 06, 2025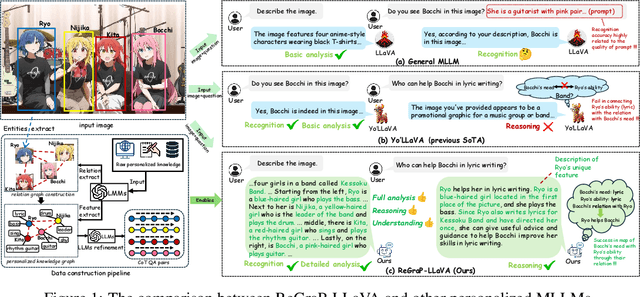

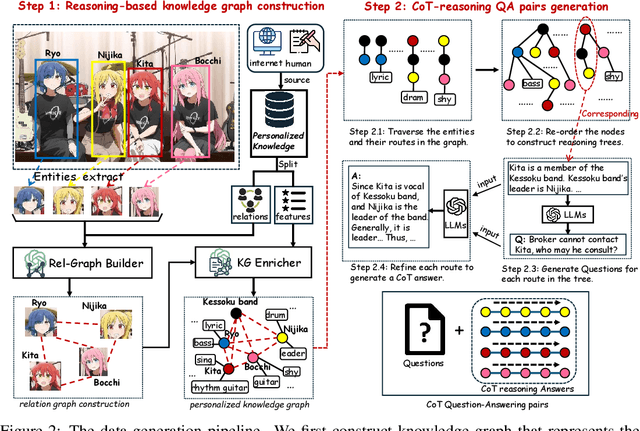
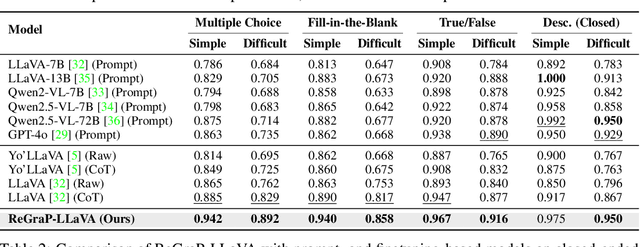
Abstract:Recent advances in personalized MLLMs enable effective capture of user-specific concepts, supporting both recognition of personalized concepts and contextual captioning. However, humans typically explore and reason over relations among objects and individuals, transcending surface-level information to achieve more personalized and contextual understanding. To this end, existing methods may face three main limitations: Their training data lacks multi-object sets in which relations among objects are learnable. Building on the limited training data, their models overlook the relations between different personalized concepts and fail to reason over them. Their experiments mainly focus on a single personalized concept, where evaluations are limited to recognition and captioning tasks. To address the limitations, we present a new dataset named ReGraP, consisting of 120 sets of personalized knowledge. Each set includes images, KGs, and CoT QA pairs derived from the KGs, enabling more structured and sophisticated reasoning pathways. We propose ReGraP-LLaVA, an MLLM trained with the corresponding KGs and CoT QA pairs, where soft and hard graph prompting methods are designed to align KGs within the model's semantic space. We establish the ReGraP Benchmark, which contains diverse task types: multiple-choice, fill-in-the-blank, True/False, and descriptive questions in both open- and closed-ended settings. The proposed benchmark is designed to evaluate the relational reasoning and knowledge-connection capability of personalized MLLMs. We conduct experiments on the proposed ReGraP-LLaVA and other competitive MLLMs. Results show that the proposed model not only learns personalized knowledge but also performs relational reasoning in responses, achieving the SoTA performance compared with the competitive methods. All the codes and datasets are released at: https://github.com/xyfyyds/ReGraP.
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
An Evidential-enhanced Tri-Branch Consistency Learning Method for Semi-supervised Medical Image Segmentation
Apr 10, 2024Abstract:Semi-supervised segmentation presents a promising approach for large-scale medical image analysis, effectively reducing annotation burdens while achieving comparable performance. This methodology holds substantial potential for streamlining the segmentation process and enhancing its feasibility within clinical settings for translational investigations. While cross-supervised training, based on distinct co-training sub-networks, has become a prevalent paradigm for this task, addressing critical issues such as predication disagreement and label-noise suppression requires further attention and progress in cross-supervised training. In this paper, we introduce an Evidential Tri-Branch Consistency learning framework (ETC-Net) for semi-supervised medical image segmentation. ETC-Net employs three branches: an evidential conservative branch, an evidential progressive branch, and an evidential fusion branch. The first two branches exhibit complementary characteristics, allowing them to address prediction diversity and enhance training stability. We also integrate uncertainty estimation from the evidential learning into cross-supervised training, mitigating the negative impact of erroneous supervision signals. Additionally, the evidential fusion branch capitalizes on the complementary attributes of the first two branches and leverages an evidence-based Dempster-Shafer fusion strategy, supervised by more reliable and accurate pseudo-labels of unlabeled data. Extensive experiments conducted on LA, Pancreas-CT, and ACDC datasets demonstrate that ETC-Net surpasses other state-of-the-art methods for semi-supervised segmentation. The code will be made available in the near future at https://github.com/Medsemiseg.
Multi Task Consistency Guided Source-Free Test-Time Domain Adaptation Medical Image Segmentation
Oct 18, 2023


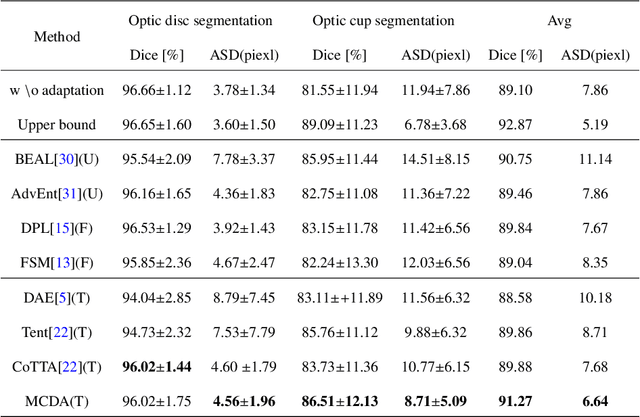
Abstract:Source-free test-time adaptation for medical image segmentation aims to enhance the adaptability of segmentation models to diverse and previously unseen test sets of the target domain, which contributes to the generalizability and robustness of medical image segmentation models without access to the source domain. Ensuring consistency between target edges and paired inputs is crucial for test-time adaptation. To improve the performance of test-time domain adaptation, we propose a multi task consistency guided source-free test-time domain adaptation medical image segmentation method which ensures the consistency of the local boundary predictions and the global prototype representation. Specifically, we introduce a local boundary consistency constraint method that explores the relationship between tissue region segmentation and tissue boundary localization tasks. Additionally, we propose a global feature consistency constraint toto enhance the intra-class compactness. We conduct extensive experiments on the segmentation of benchmark fundus images. Compared to prediction directly by the source domain model, the segmentation Dice score is improved by 6.27\% and 0.96\% in RIM-ONE-r3 and Drishti GS datasets, respectively. Additionally, the results of experiments demonstrate that our proposed method outperforms existing competitive domain adaptation segmentation algorithms.
Topology-inspired Cross-domain Network for Developmental Cervical Stenosis Quantification
Sep 18, 2023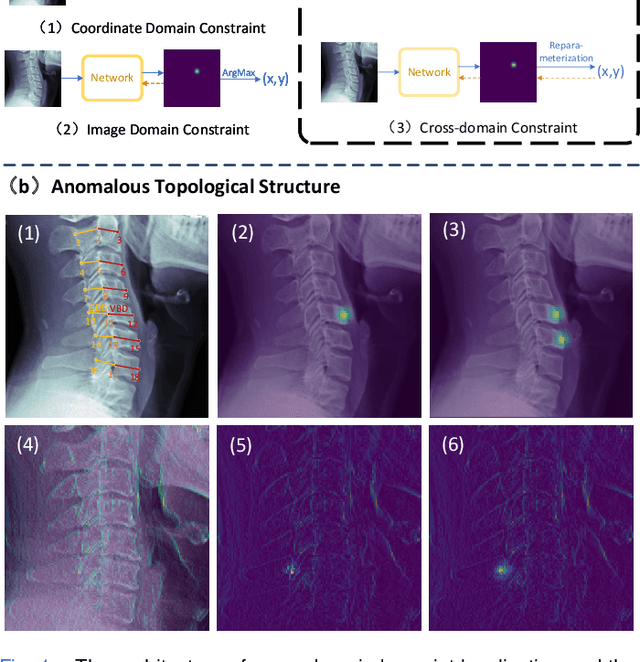
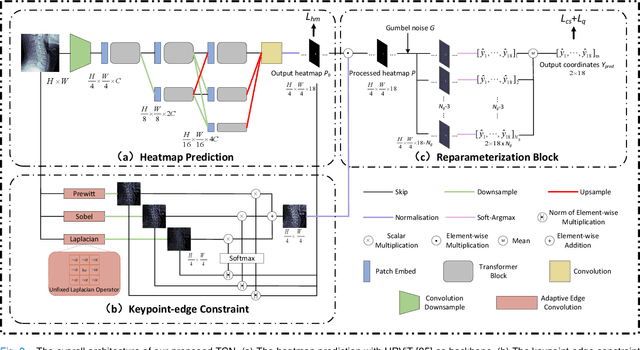
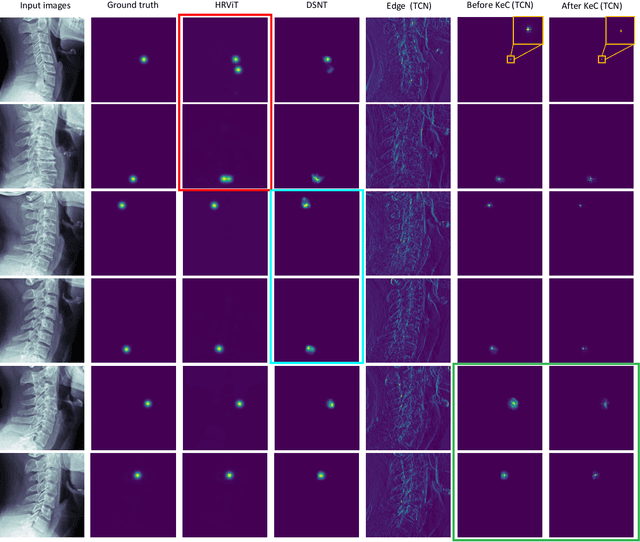
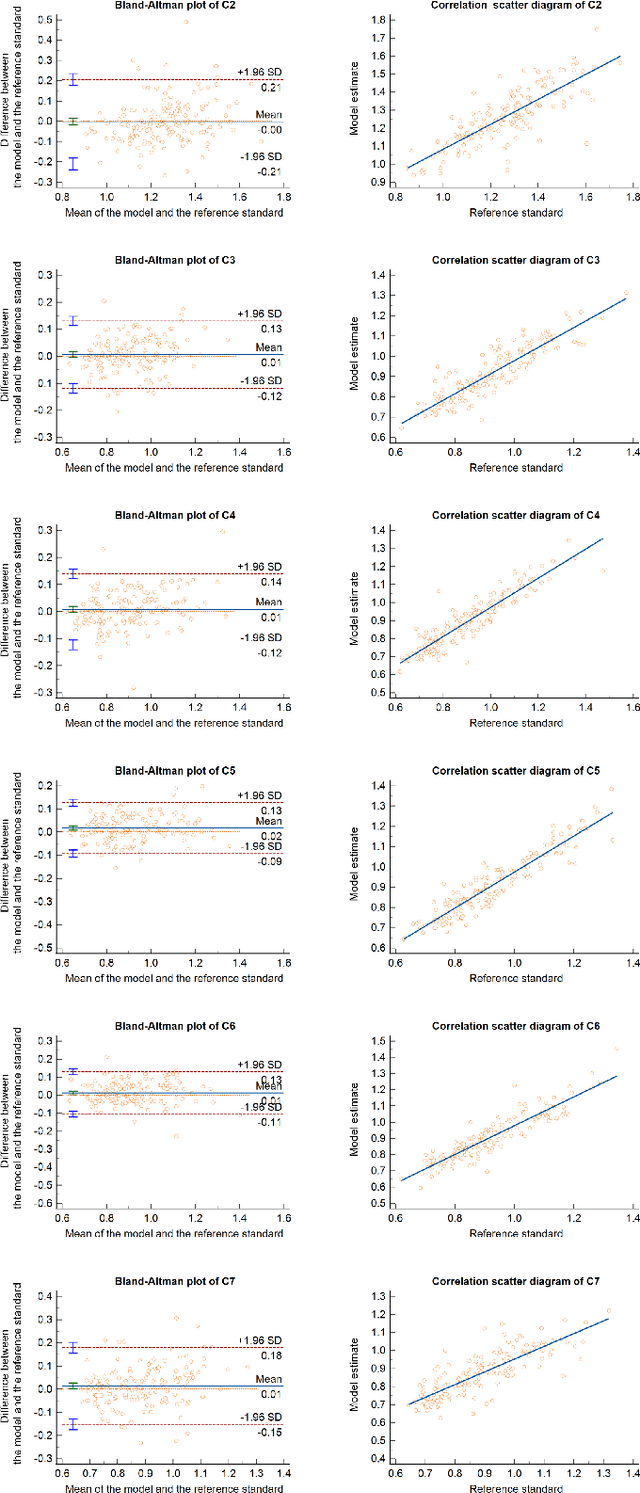
Abstract:Developmental Canal Stenosis (DCS) quantification is crucial in cervical spondylosis screening. Compared with quantifying DCS manually, a more efficient and time-saving manner is provided by deep keypoint localization networks, which can be implemented in either the coordinate or the image domain. However, the vertebral visualization features often lead to abnormal topological structures during keypoint localization, including keypoint distortion with edges and weakly connected structures, which cannot be fully suppressed in either the coordinate or image domain alone. To overcome this limitation, a keypoint-edge and a reparameterization modules are utilized to restrict these abnormal structures in a cross-domain manner. The keypoint-edge constraint module restricts the keypoints on the edges of vertebrae, which ensures that the distribution pattern of keypoint coordinates is consistent with those for DCS quantification. And the reparameterization module constrains the weakly connected structures in image-domain heatmaps with coordinates combined. Moreover, the cross-domain network improves spatial generalization by utilizing heatmaps and incorporating coordinates for accurate localization, which avoids the trade-off between these two properties in an individual domain. Comprehensive results of distinct quantification tasks show the superiority and generability of the proposed Topology-inspired Cross-domain Network (TCN) compared with other competing localization methods.
Cross-supervised Dual Classifiers for Semi-supervised Medical Image Segmentation
May 25, 2023Abstract:Semi-supervised medical image segmentation offers a promising solution for large-scale medical image analysis by significantly reducing the annotation burden while achieving comparable performance. Employing this method exhibits a high degree of potential for optimizing the segmentation process and increasing its feasibility in clinical settings during translational investigations. Recently, cross-supervised training based on different co-training sub-networks has become a standard paradigm for this task. Still, the critical issues of sub-network disagreement and label-noise suppression require further attention and progress in cross-supervised training. This paper proposes a cross-supervised learning framework based on dual classifiers (DC-Net), including an evidential classifier and a vanilla classifier. The two classifiers exhibit complementary characteristics, enabling them to handle disagreement effectively and generate more robust and accurate pseudo-labels for unlabeled data. We also incorporate the uncertainty estimation from the evidential classifier into cross-supervised training to alleviate the negative effect of the error supervision signal. The extensive experiments on LA and Pancreas-CT dataset illustrate that DC-Net outperforms other state-of-the-art methods for semi-supervised segmentation. The code will be released soon.
Self-aware and Cross-sample Prototypical Learning for Semi-supervised Medical Image Segmentation
May 25, 2023



Abstract:Consistency learning plays a crucial role in semi-supervised medical image segmentation as it enables the effective utilization of limited annotated data while leveraging the abundance of unannotated data. The effectiveness and efficiency of consistency learning are challenged by prediction diversity and training stability, which are often overlooked by existing studies. Meanwhile, the limited quantity of labeled data for training often proves inadequate for formulating intra-class compactness and inter-class discrepancy of pseudo labels. To address these issues, we propose a self-aware and cross-sample prototypical learning method (SCP-Net) to enhance the diversity of prediction in consistency learning by utilizing a broader range of semantic information derived from multiple inputs. Furthermore, we introduce a self-aware consistency learning method that exploits unlabeled data to improve the compactness of pseudo labels within each class. Moreover, a dual loss re-weighting method is integrated into the cross-sample prototypical consistency learning method to improve the reliability and stability of our model. Extensive experiments on ACDC dataset and PROMISE12 dataset validate that SCP-Net outperforms other state-of-the-art semi-supervised segmentation methods and achieves significant performance gains compared to the limited supervised training. Our code will come soon.
Position-Aware Relation Learning for RGB-Thermal Salient Object Detection
Sep 21, 2022
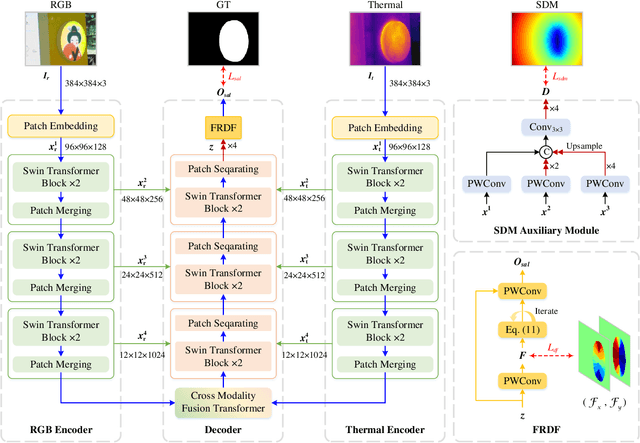
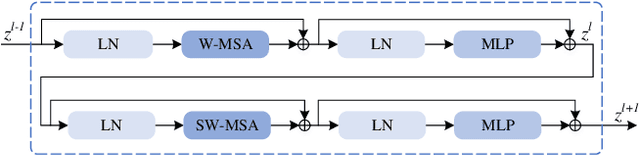

Abstract:RGB-Thermal salient object detection (SOD) combines two spectra to segment visually conspicuous regions in images. Most existing methods use boundary maps to learn the sharp boundary. These methods ignore the interactions between isolated boundary pixels and other confident pixels, leading to sub-optimal performance. To address this problem,we propose a position-aware relation learning network (PRLNet) for RGB-T SOD based on swin transformer. PRLNet explores the distance and direction relationships between pixels to strengthen intra-class compactness and inter-class separation, generating salient object masks with clear boundaries and homogeneous regions. Specifically, we develop a novel signed distance map auxiliary module (SDMAM) to improve encoder feature representation, which takes into account the distance relation of different pixels in boundary neighborhoods. Then, we design a feature refinement approach with directional field (FRDF), which rectifies features of boundary neighborhood by exploiting the features inside salient objects. FRDF utilizes the directional information between object pixels to effectively enhance the intra-class compactness of salient regions. In addition, we constitute a pure transformer encoder-decoder network to enhance multispectral feature representation for RGB-T SOD. Finally, we conduct quantitative and qualitative experiments on three public benchmark datasets.The results demonstrate that our proposed method outperforms the state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge