Pietro Cerveri
Politecnico di Milano, Università di Pavia
Generalist Models in Medical Image Segmentation: A Survey and Performance Comparison with Task-Specific Approaches
Jun 12, 2025



Abstract:Following the successful paradigm shift of large language models, leveraging pre-training on a massive corpus of data and fine-tuning on different downstream tasks, generalist models have made their foray into computer vision. The introduction of Segment Anything Model (SAM) set a milestone on segmentation of natural images, inspiring the design of a multitude of architectures for medical image segmentation. In this survey we offer a comprehensive and in-depth investigation on generalist models for medical image segmentation. We start with an introduction on the fundamentals concepts underpinning their development. Then, we provide a taxonomy on the different declinations of SAM in terms of zero-shot, few-shot, fine-tuning, adapters, on the recent SAM 2, on other innovative models trained on images alone, and others trained on both text and images. We thoroughly analyze their performances at the level of both primary research and best-in-literature, followed by a rigorous comparison with the state-of-the-art task-specific models. We emphasize the need to address challenges in terms of compliance with regulatory frameworks, privacy and security laws, budget, and trustworthy artificial intelligence (AI). Finally, we share our perspective on future directions concerning synthetic data, early fusion, lessons learnt from generalist models in natural language processing, agentic AI and physical AI, and clinical translation.
MiniGPT-Pancreas: Multimodal Large Language Model for Pancreas Cancer Classification and Detection
Dec 20, 2024

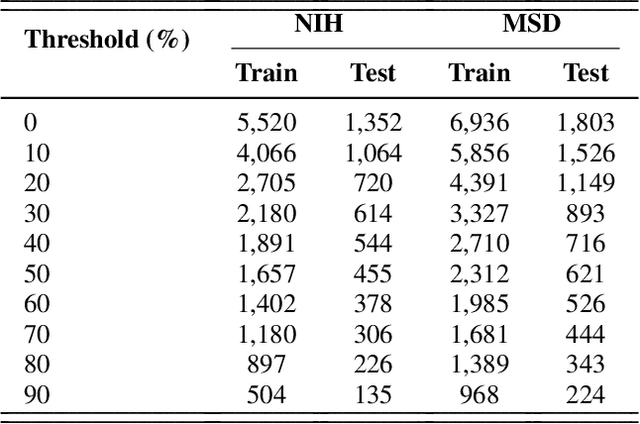
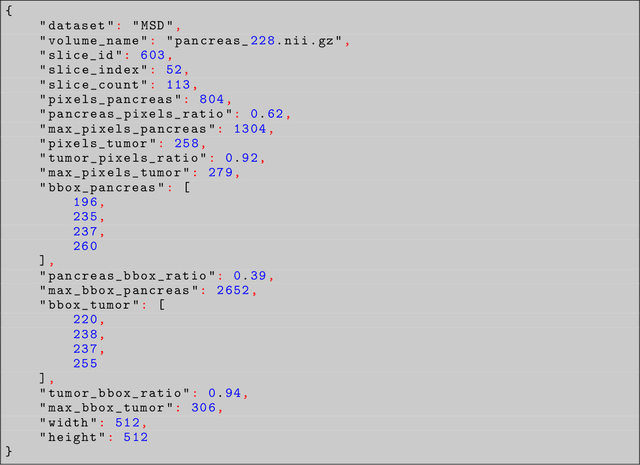
Abstract:Problem: Pancreas radiological imaging is challenging due to the small size, blurred boundaries, and variability of shape and position of the organ among patients. Goal: In this work we present MiniGPT-Pancreas, a Multimodal Large Language Model (MLLM), as an interactive chatbot to support clinicians in pancreas cancer diagnosis by integrating visual and textual information. Methods: MiniGPT-v2, a general-purpose MLLM, was fine-tuned in a cascaded way for pancreas detection, tumor classification, and tumor detection with multimodal prompts combining questions and computed tomography scans from the National Institute of Health (NIH), and Medical Segmentation Decathlon (MSD) datasets. The AbdomenCT-1k dataset was used to detect the liver, spleen, kidney, and pancreas. Results: MiniGPT-Pancreas achieved an Intersection over Union (IoU) of 0.595 and 0.550 for the detection of pancreas on NIH and MSD datasets, respectively. For the pancreas cancer classification task on the MSD dataset, accuracy, precision, and recall were 0.876, 0.874, and 0.878, respectively. When evaluating MiniGPT-Pancreas on the AbdomenCT-1k dataset for multi-organ detection, the IoU was 0.8399 for the liver, 0.722 for the kidney, 0.705 for the spleen, and 0.497 for the pancreas. For the pancreas tumor detection task, the IoU score was 0.168 on the MSD dataset. Conclusions: MiniGPT-Pancreas represents a promising solution to support clinicians in the classification of pancreas images with pancreas tumors. Future research is needed to improve the score on the detection task, especially for pancreas tumors.
Optimized two-stage AI-based Neural Decoding for Enhanced Visual Stimulus Reconstruction from fMRI Data
Dec 17, 2024



Abstract:AI-based neural decoding reconstructs visual perception by leveraging generative models to map brain activity, measured through functional MRI (fMRI), into latent hierarchical representations. Traditionally, ridge linear models transform fMRI into a latent space, which is then decoded using latent diffusion models (LDM) via a pre-trained variational autoencoder (VAE). Due to the complexity and noisiness of fMRI data, newer approaches split the reconstruction into two sequential steps, the first one providing a rough visual approximation, the second on improving the stimulus prediction via LDM endowed by CLIP embeddings. This work proposes a non-linear deep network to improve fMRI latent space representation, optimizing the dimensionality alike. Experiments on the Natural Scenes Dataset showed that the proposed architecture improved the structural similarity of the reconstructed image by about 2\% with respect to the state-of-the-art model, based on ridge linear transform. The reconstructed image's semantics improved by about 4\%, measured by perceptual similarity, with respect to the state-of-the-art. The noise sensitivity analysis of the LDM showed that the role of the first stage was fundamental to predict the stimulus featuring high structural similarity. Conversely, providing a large noise stimulus affected less the semantics of the predicted stimulus, while the structural similarity between the ground truth and predicted stimulus was very poor. The findings underscore the importance of leveraging non-linear relationships between BOLD signal and the latent representation and two-stage generative AI for optimizing the fidelity of reconstructed visual stimuli from noisy fMRI data.
Cascade learning in multi-task encoder-decoder networks for concurrent bone segmentation and glenohumeral joint assessment in shoulder CT scans
Oct 16, 2024



Abstract:Osteoarthritis is a degenerative condition affecting bones and cartilage, often leading to osteophyte formation, bone density loss, and joint space narrowing. Treatment options to restore normal joint function vary depending on the severity of the condition. This work introduces an innovative deep-learning framework processing shoulder CT scans. It features the semantic segmentation of the proximal humerus and scapula, the 3D reconstruction of bone surfaces, the identification of the glenohumeral (GH) joint region, and the staging of three common osteoarthritic-related pathologies: osteophyte formation (OS), GH space reduction (JS), and humeroscapular alignment (HSA). The pipeline comprises two cascaded CNN architectures: 3D CEL-UNet for segmentation and 3D Arthro-Net for threefold classification. A retrospective dataset of 571 CT scans featuring patients with various degrees of GH osteoarthritic-related pathologies was used to train, validate, and test the pipeline. Root mean squared error and Hausdorff distance median values for 3D reconstruction were 0.22mm and 1.48mm for the humerus and 0.24mm and 1.48mm for the scapula, outperforming state-of-the-art architectures and making it potentially suitable for a PSI-based shoulder arthroplasty preoperative plan context. The classification accuracy for OS, JS, and HSA consistently reached around 90% across all three categories. The computational time for the inference pipeline was less than 15s, showcasing the framework's efficiency and compatibility with orthopedic radiology practice. The outcomes represent a promising advancement toward the medical translation of artificial intelligence tools. This progress aims to streamline the preoperative planning pipeline delivering high-quality bone surfaces and supporting surgeons in selecting the most suitable surgical approach according to the unique patient joint conditions.
Deep Learning for Pancreas Segmentation: a Systematic Review
Jul 23, 2024Abstract:Pancreas segmentation has been traditionally challenging due to its small size in computed tomography abdominal volumes, high variability of shape and positions among patients, and blurred boundaries due to low contrast between the pancreas and surrounding organs. Many deep learning models for pancreas segmentation have been proposed in the past few years. We present a thorough systematic review based on the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) statement. The literature search was conducted on PubMed, Web of Science, Scopus, and IEEE Xplore on original studies published in peer-reviewed journals from 2013 to 2023. Overall, 130 studies were retrieved. We initially provided an overview of the technical background of the most common network architectures and publicly available datasets. Then, the analysis of the studies combining visual presentation in tabular form and text description was reported. The tables grouped the studies specifying the application, dataset size, design (model architecture, learning strategy, and loss function), results, and main contributions. We first analyzed the studies focusing on parenchyma segmentation using coarse-to-fine approaches, multi-organ segmentation, semi-supervised learning, and unsupervised learning, followed by those studies on generalization to other datasets and those concerning the design of new loss functions. Then, we analyzed the studies on segmentation of tumors, cysts, and inflammation reporting multi-stage methods, semi-supervised learning, generalization to other datasets, and design of new loss functions. Finally, we provided a critical discussion on the subject based on the published evidence underlining current issues that need to be addressed before clinical translation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge