Yunlong Zhao
Speech-Aware Long Context Pruning and Integration for Contextualized Automatic Speech Recognition
Nov 14, 2025Abstract:Automatic speech recognition (ASR) systems have achieved remarkable performance in common conditions but often struggle to leverage long-context information in contextualized scenarios that require domain-specific knowledge, such as conference presentations. This challenge arises primarily due to constrained model context windows and the sparsity of relevant information within extensive contextual noise. To solve this, we propose the SAP$^{2}$ method, a novel framework that dynamically prunes and integrates relevant contextual keywords in two stages. Specifically, each stage leverages our proposed Speech-Driven Attention-based Pooling mechanism, enabling efficient compression of context embeddings while preserving speech-salient information. Experimental results demonstrate state-of-the-art performance of SAP$^{2}$ on the SlideSpeech and LibriSpeech datasets, achieving word error rates (WER) of 7.71% and 1.12%, respectively. On SlideSpeech, our method notably reduces biased keyword error rates (B-WER) by 41.1% compared to non-contextual baselines. SAP$^{2}$ also exhibits robust scalability, consistently maintaining performance under extensive contextual input conditions on both datasets.
PRNet: Original Information Is All You Have
Oct 10, 2025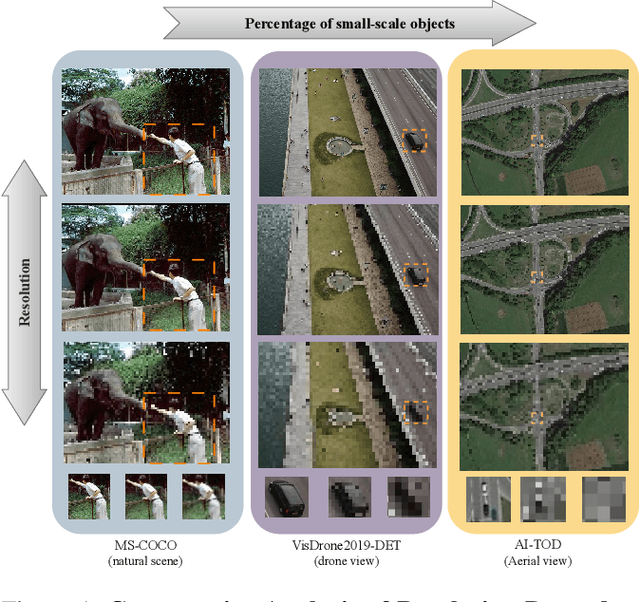
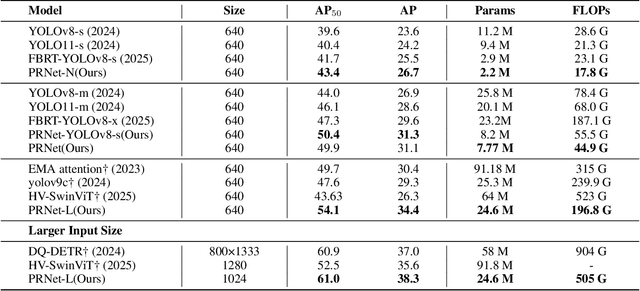
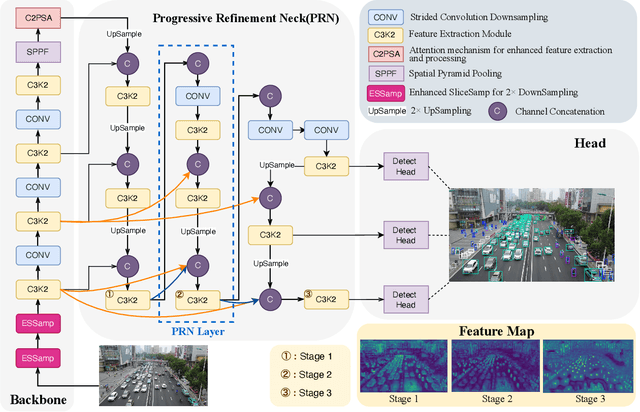
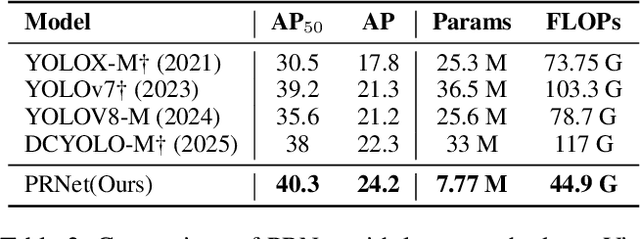
Abstract:Small object detection in aerial images suffers from severe information degradation during feature extraction due to limited pixel representations, where shallow spatial details fail to align effectively with semantic information, leading to frequent misses and false positives. Existing FPN-based methods attempt to mitigate these losses through post-processing enhancements, but the reconstructed details often deviate from the original image information, impeding their fusion with semantic content. To address this limitation, we propose PRNet, a real-time detection framework that prioritizes the preservation and efficient utilization of primitive shallow spatial features to enhance small object representations. PRNet achieves this via two modules:the Progressive Refinement Neck (PRN) for spatial-semantic alignment through backbone reuse and iterative refinement, and the Enhanced SliceSamp (ESSamp) for preserving shallow information during downsampling via optimized rearrangement and convolution. Extensive experiments on the VisDrone, AI-TOD, and UAVDT datasets demonstrate that PRNet outperforms state-of-the-art methods under comparable computational constraints, achieving superior accuracy-efficiency trade-offs.
DPMT: Dual Process Multi-scale Theory of Mind Framework for Real-time Human-AI Collaboration
Jul 18, 2025Abstract:Real-time human-artificial intelligence (AI) collaboration is crucial yet challenging, especially when AI agents must adapt to diverse and unseen human behaviors in dynamic scenarios. Existing large language model (LLM) agents often fail to accurately model the complex human mental characteristics such as domain intentions, especially in the absence of direct communication. To address this limitation, we propose a novel dual process multi-scale theory of mind (DPMT) framework, drawing inspiration from cognitive science dual process theory. Our DPMT framework incorporates a multi-scale theory of mind (ToM) module to facilitate robust human partner modeling through mental characteristic reasoning. Experimental results demonstrate that DPMT significantly enhances human-AI collaboration, and ablation studies further validate the contributions of our multi-scale ToM in the slow system.
LVC: A Lightweight Compression Framework for Enhancing VLMs in Long Video Understanding
Apr 09, 2025Abstract:Long video understanding is a complex task that requires both spatial detail and temporal awareness. While Vision-Language Models (VLMs) obtain frame-level understanding capabilities through multi-frame input, they suffer from information loss due to the sparse sampling strategy. In contrast, Video Large Language Models (Video-LLMs) capture temporal relationships within visual features but are limited by the scarcity of high-quality video-text datasets. To transfer long video understanding capabilities to VLMs with minimal data and computational cost, we propose Lightweight Video Compression (LVC), a novel method featuring the Query-Attention Video Compression mechanism, which effectively tackles the sparse sampling problem in VLMs. By training only the alignment layer with 10k short video-text pairs, LVC significantly enhances the temporal reasoning abilities of VLMs. Extensive experiments show that LVC provides consistent performance improvements across various models, including the InternVL2 series and Phi-3.5-Vision. Notably, the InternVL2-40B-LVC achieves scores of 68.2 and 65.9 on the long video understanding benchmarks MLVU and Video-MME, respectively, with relative improvements of 14.6% and 7.7%. The enhanced models and code will be publicly available soon.
GridShow: Omni Visual Generation
Dec 17, 2024Abstract:In this paper, we introduce GRID, a novel paradigm that reframes a broad range of visual generation tasks as the problem of arranging grids, akin to film strips. At its core, GRID transforms temporal sequences into grid layouts, enabling image generation models to process visual sequences holistically. To achieve both layout consistency and motion coherence, we develop a parallel flow-matching training strategy that combines layout matching and temporal losses, guided by a coarse-to-fine schedule that evolves from basic layouts to precise motion control. Our approach demonstrates remarkable efficiency, achieving up to 35 faster inference speeds while using 1/1000 of the computational resources compared to specialized models. Extensive experiments show that GRID exhibits exceptional versatility across diverse visual generation tasks, from Text-to-Video to 3D Editing, while maintaining its foundational image generation capabilities. This dual strength in both expanded applications and preserved core competencies establishes GRID as an efficient and versatile omni-solution for visual generation.
MetaDD: Boosting Dataset Distillation with Neural Network Architecture-Invariant Generalization
Oct 07, 2024



Abstract:Dataset distillation (DD) entails creating a refined, compact distilled dataset from a large-scale dataset to facilitate efficient training. A significant challenge in DD is the dependency between the distilled dataset and the neural network (NN) architecture used. Training a different NN architecture with a distilled dataset distilled using a specific architecture often results in diminished trainning performance for other architectures. This paper introduces MetaDD, designed to enhance the generalizability of DD across various NN architectures. Specifically, MetaDD partitions distilled data into meta features (i.e., the data's common characteristics that remain consistent across different NN architectures) and heterogeneous features (i.e., the data's unique feature to each NN architecture). Then, MetaDD employs an architecture-invariant loss function for multi-architecture feature alignment, which increases meta features and reduces heterogeneous features in distilled data. As a low-memory consumption component, MetaDD can be seamlessly integrated into any DD methodology. Experimental results demonstrate that MetaDD significantly improves performance across various DD methods. On the Distilled Tiny-Imagenet with Sre2L (50 IPC), MetaDD achieves cross-architecture NN accuracy of up to 30.1\%, surpassing the second-best method (GLaD) by 1.7\%.
Generative Enzyme Design Guided by Functionally Important Sites and Small-Molecule Substrates
May 13, 2024Abstract:Enzymes are genetically encoded biocatalysts capable of accelerating chemical reactions. How can we automatically design functional enzymes? In this paper, we propose EnzyGen, an approach to learn a unified model to design enzymes across all functional families. Our key idea is to generate an enzyme's amino acid sequence and their three-dimensional (3D) coordinates based on functionally important sites and substrates corresponding to a desired catalytic function. These sites are automatically mined from enzyme databases. EnzyGen consists of a novel interleaving network of attention and neighborhood equivariant layers, which captures both long-range correlation in an entire protein sequence and local influence from nearest amino acids in 3D space. To learn the generative model, we devise a joint training objective, including a sequence generation loss, a position prediction loss and an enzyme-substrate interaction loss. We further construct EnzyBench, a dataset with 3157 enzyme families, covering all available enzymes within the protein data bank (PDB). Experimental results show that our EnzyGen consistently achieves the best performance across all 323 testing families, surpassing the best baseline by 10.79% in terms of substrate binding affinity. These findings demonstrate EnzyGen's superior capability in designing well-folded and effective enzymes binding to specific substrates with high affinities.
Generating Games via LLMs: An Investigation with Video Game Description Language
Apr 11, 2024



Abstract:Recently, the emergence of large language models (LLMs) has unlocked new opportunities for procedural content generation. However, recent attempts mainly focus on level generation for specific games with defined game rules such as Super Mario Bros. and Zelda. This paper investigates the game generation via LLMs. Based on video game description language, this paper proposes an LLM-based framework to generate game rules and levels simultaneously. Experiments demonstrate how the framework works with prompts considering different combinations of context. Our findings extend the current applications of LLMs and offer new insights for generating new games in the area of procedural content generation.
Functional Geometry Guided Protein Sequence and Backbone Structure Co-Design
Oct 09, 2023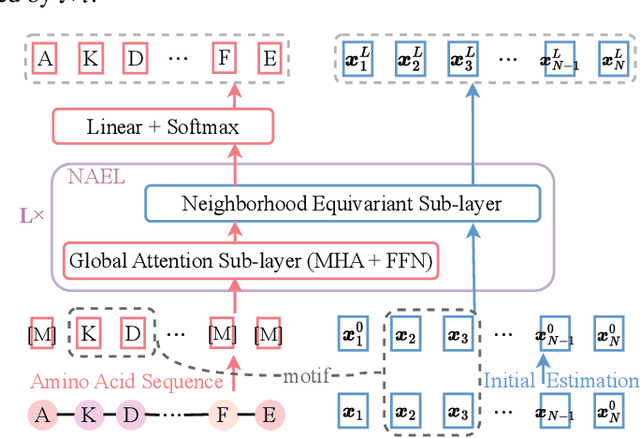
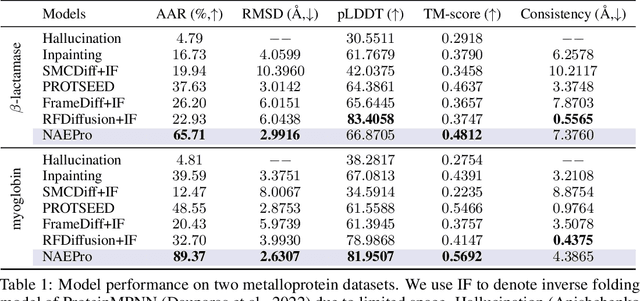
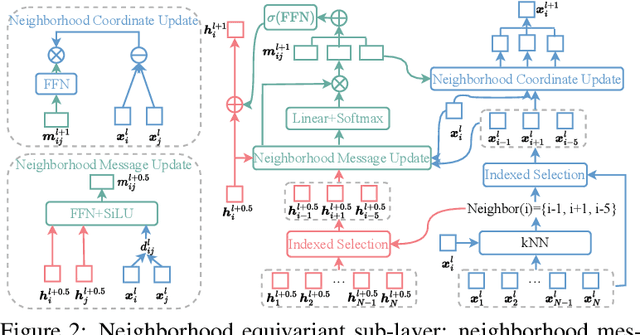
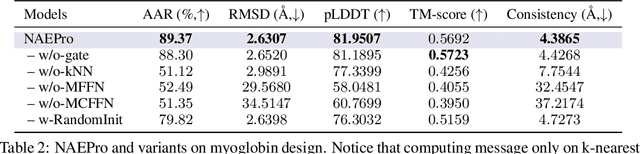
Abstract:Proteins are macromolecules responsible for essential functions in almost all living organisms. Designing reasonable proteins with desired functions is crucial. A protein's sequence and structure are strongly correlated and they together determine its function. In this paper, we propose NAEPro, a model to jointly design Protein sequence and structure based on automatically detected functional sites. NAEPro is powered by an interleaving network of attention and equivariant layers, which can capture global correlation in a whole sequence and local influence from nearest amino acids in three dimensional (3D) space. Such an architecture facilitates effective yet economic message passing at two levels. We evaluate our model and several strong baselines on two protein datasets, $\beta$-lactamase and myoglobin. Experimental results show that our model consistently achieves the highest amino acid recovery rate, TM-score, and the lowest RMSD among all competitors. These findings prove the capability of our model to design protein sequences and structures that closely resemble their natural counterparts. Furthermore, in-depth analysis further confirms our model's ability to generate highly effective proteins capable of binding to their target metallocofactors. We provide code, data and models in Github.
Joint Design of Protein Sequence and Structure based on Motifs
Oct 04, 2023Abstract:Designing novel proteins with desired functions is crucial in biology and chemistry. However, most existing work focus on protein sequence design, leaving protein sequence and structure co-design underexplored. In this paper, we propose GeoPro, a method to design protein backbone structure and sequence jointly. Our motivation is that protein sequence and its backbone structure constrain each other, and thus joint design of both can not only avoid nonfolding and misfolding but also produce more diverse candidates with desired functions. To this end, GeoPro is powered by an equivariant encoder for three-dimensional (3D) backbone structure and a protein sequence decoder guided by 3D geometry. Experimental results on two biologically significant metalloprotein datasets, including $\beta$-lactamases and myoglobins, show that our proposed GeoPro outperforms several strong baselines on most metrics. Remarkably, our method discovers novel $\beta$-lactamases and myoglobins which are not present in protein data bank (PDB) and UniProt. These proteins exhibit stable folding and active site environments reminiscent of those of natural proteins, demonstrating their excellent potential to be biologically functional.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge