Yufei Song
UnlearnShield: Shielding Forgotten Privacy against Unlearning Inversion
Jan 28, 2026Abstract:Machine unlearning is an emerging technique that aims to remove the influence of specific data from trained models, thereby enhancing privacy protection. However, recent research has uncovered critical privacy vulnerabilities, showing that adversaries can exploit unlearning inversion to reconstruct data that was intended to be erased. Despite the severity of this threat, dedicated defenses remain lacking. To address this gap, we propose UnlearnShield, the first defense specifically tailored to counter unlearning inversion. UnlearnShield introduces directional perturbations in the cosine representation space and regulates them through a constraint module to jointly preserve model accuracy and forgetting efficacy, thereby reducing inversion risk while maintaining utility. Experiments demonstrate that it achieves a good trade-off among privacy protection, accuracy, and forgetting.
Erosion Attack for Adversarial Training to Enhance Semantic Segmentation Robustness
Jan 21, 2026Abstract:Existing segmentation models exhibit significant vulnerability to adversarial attacks.To improve robustness, adversarial training incorporates adversarial examples into model training. However, existing attack methods consider only global semantic information and ignore contextual semantic relationships within the samples, limiting the effectiveness of adversarial training. To address this issue, we propose EroSeg-AT, a vulnerability-aware adversarial training framework that leverages EroSeg to generate adversarial examples. EroSeg first selects sensitive pixels based on pixel-level confidence and then progressively propagates perturbations to higher-confidence pixels, effectively disrupting the semantic consistency of the samples. Experimental results show that, compared to existing methods, our approach significantly improves attack effectiveness and enhances model robustness under adversarial training.
NumbOD: A Spatial-Frequency Fusion Attack Against Object Detectors
Dec 22, 2024Abstract:With the advancement of deep learning, object detectors (ODs) with various architectures have achieved significant success in complex scenarios like autonomous driving. Previous adversarial attacks against ODs have been focused on designing customized attacks targeting their specific structures (e.g., NMS and RPN), yielding some results but simultaneously constraining their scalability. Moreover, most efforts against ODs stem from image-level attacks originally designed for classification tasks, resulting in redundant computations and disturbances in object-irrelevant areas (e.g., background). Consequently, how to design a model-agnostic efficient attack to comprehensively evaluate the vulnerabilities of ODs remains challenging and unresolved. In this paper, we propose NumbOD, a brand-new spatial-frequency fusion attack against various ODs, aimed at disrupting object detection within images. We directly leverage the features output by the OD without relying on its internal structures to craft adversarial examples. Specifically, we first design a dual-track attack target selection strategy to select high-quality bounding boxes from OD outputs for targeting. Subsequently, we employ directional perturbations to shift and compress predicted boxes and change classification results to deceive ODs. Additionally, we focus on manipulating the high-frequency components of images to confuse ODs' attention on critical objects, thereby enhancing the attack efficiency. Our extensive experiments on nine ODs and two datasets show that NumbOD achieves powerful attack performance and high stealthiness.
PB-UAP: Hybrid Universal Adversarial Attack For Image Segmentation
Dec 21, 2024



Abstract:With the rapid advancement of deep learning, the model robustness has become a significant research hotspot, \ie, adversarial attacks on deep neural networks. Existing works primarily focus on image classification tasks, aiming to alter the model's predicted labels. Due to the output complexity and deeper network architectures, research on adversarial examples for segmentation models is still limited, particularly for universal adversarial perturbations. In this paper, we propose a novel universal adversarial attack method designed for segmentation models, which includes dual feature separation and low-frequency scattering modules. The two modules guide the training of adversarial examples in the pixel and frequency space, respectively. Experiments demonstrate that our method achieves high attack success rates surpassing the state-of-the-art methods, and exhibits strong transferability across different models.
Trimming Down Large Spiking Vision Transformers via Heterogeneous Quantization Search
Dec 07, 2024



Abstract:Spiking Neural Networks (SNNs) are amenable to deployment on edge devices and neuromorphic hardware due to their lower dissipation. Recently, SNN-based transformers have garnered significant interest, incorporating attention mechanisms akin to their counterparts in Artificial Neural Networks (ANNs) while demonstrating excellent performance. However, deploying large spiking transformer models on resource-constrained edge devices such as mobile phones, still poses significant challenges resulted from the high computational demands of large uncompressed high-precision models. In this work, we introduce a novel heterogeneous quantization method for compressing spiking transformers through layer-wise quantization. Our approach optimizes the quantization of each layer using one of two distinct quantization schemes, i.e., uniform or power-of-two quantification, with mixed bit resolutions. Our heterogeneous quantization demonstrates the feasibility of maintaining high performance for spiking transformers while utilizing an average effective resolution of 3.14-3.67 bits with less than a 1% accuracy drop on DVS Gesture and CIFAR10-DVS datasets. It attains a model compression rate of 8.71x-10.19x for standard floating-point spiking transformers. Moreover, the proposed approach achieves a significant energy reduction of 5.69x, 8.72x, and 10.2x while maintaining high accuracy levels of 85.3%, 97.57%, and 80.4% on N-Caltech101, DVS-Gesture, and CIFAR10-DVS datasets, respectively.
DarkSAM: Fooling Segment Anything Model to Segment Nothing
Sep 26, 2024

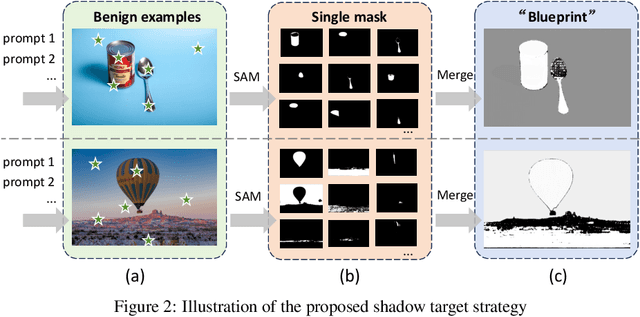
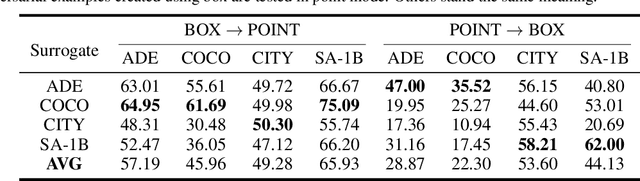
Abstract:Segment Anything Model (SAM) has recently gained much attention for its outstanding generalization to unseen data and tasks. Despite its promising prospect, the vulnerabilities of SAM, especially to universal adversarial perturbation (UAP) have not been thoroughly investigated yet. In this paper, we propose DarkSAM, the first prompt-free universal attack framework against SAM, including a semantic decoupling-based spatial attack and a texture distortion-based frequency attack. We first divide the output of SAM into foreground and background. Then, we design a shadow target strategy to obtain the semantic blueprint of the image as the attack target. DarkSAM is dedicated to fooling SAM by extracting and destroying crucial object features from images in both spatial and frequency domains. In the spatial domain, we disrupt the semantics of both the foreground and background in the image to confuse SAM. In the frequency domain, we further enhance the attack effectiveness by distorting the high-frequency components (i.e., texture information) of the image. Consequently, with a single UAP, DarkSAM renders SAM incapable of segmenting objects across diverse images with varying prompts. Experimental results on four datasets for SAM and its two variant models demonstrate the powerful attack capability and transferability of DarkSAM.
Joint Design of Protein Sequence and Structure based on Motifs
Oct 04, 2023Abstract:Designing novel proteins with desired functions is crucial in biology and chemistry. However, most existing work focus on protein sequence design, leaving protein sequence and structure co-design underexplored. In this paper, we propose GeoPro, a method to design protein backbone structure and sequence jointly. Our motivation is that protein sequence and its backbone structure constrain each other, and thus joint design of both can not only avoid nonfolding and misfolding but also produce more diverse candidates with desired functions. To this end, GeoPro is powered by an equivariant encoder for three-dimensional (3D) backbone structure and a protein sequence decoder guided by 3D geometry. Experimental results on two biologically significant metalloprotein datasets, including $\beta$-lactamases and myoglobins, show that our proposed GeoPro outperforms several strong baselines on most metrics. Remarkably, our method discovers novel $\beta$-lactamases and myoglobins which are not present in protein data bank (PDB) and UniProt. These proteins exhibit stable folding and active site environments reminiscent of those of natural proteins, demonstrating their excellent potential to be biologically functional.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge