Weihao Gao
Enabling Real-Time Colonoscopic Polyp Segmentation on Commodity CPUs via Ultra-Lightweight Architecture
Feb 04, 2026Abstract:Early detection of colorectal cancer hinges on real-time, accurate polyp identification and resection. Yet current high-precision segmentation models rely on GPUs, making them impractical to deploy in primary hospitals, mobile endoscopy units, or capsule robots. To bridge this gap, we present the UltraSeg family, operating in an extreme-compression regime (<0.3 M parameters). UltraSeg-108K (0.108 M parameters) is optimized for single-center data, while UltraSeg-130K (0.13 M parameters) generalizes to multi-center, multi-modal images. By jointly optimizing encoder-decoder widths, incorporating constrained dilated convolutions to enlarge receptive fields, and integrating a cross-layer lightweight fusion module, the models achieve 90 FPS on a single CPU core without sacrificing accuracy. Evaluated on seven public datasets, UltraSeg retains >94% of the Dice score of a 31 M-parameter U-Net while utilizing only 0.4% of its parameters, establishing a strong, clinically viable baseline for the extreme-compression domain and offering an immediately deployable solution for resource-constrained settings. This work provides not only a CPU-native solution for colonoscopy but also a reproducible blueprint for broader minimally invasive surgical vision applications. Source code is publicly available to ensure reproducibility and facilitate future benchmarking.
Seed-Prover 1.5: Mastering Undergraduate-Level Theorem Proving via Learning from Experience
Dec 19, 2025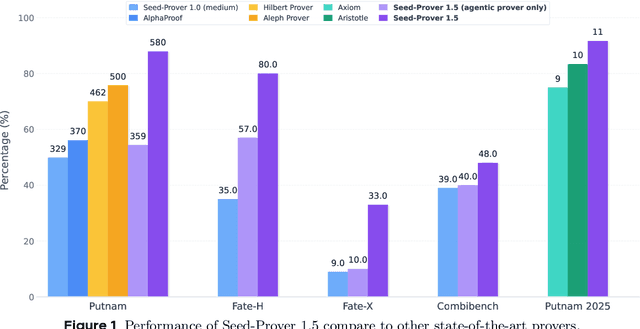

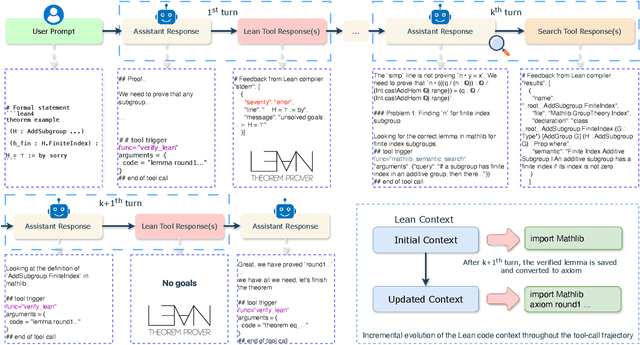
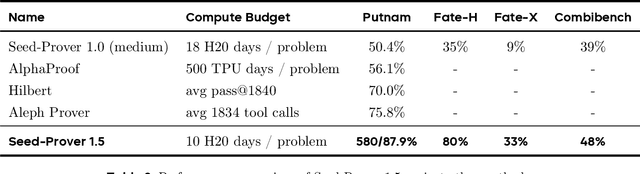
Abstract:Large language models have recently made significant progress to generate rigorous mathematical proofs. In contrast, utilizing LLMs for theorem proving in formal languages (such as Lean) remains challenging and computationally expensive, particularly when addressing problems at the undergraduate level and beyond. In this work, we present \textbf{Seed-Prover 1.5}, a formal theorem-proving model trained via large-scale agentic reinforcement learning, alongside an efficient test-time scaling (TTS) workflow. Through extensive interactions with Lean and other tools, the model continuously accumulates experience during the RL process, substantially enhancing the capability and efficiency of formal theorem proving. Furthermore, leveraging recent advancements in natural language proving, our TTS workflow efficiently bridges the gap between natural and formal languages. Compared to state-of-the-art methods, Seed-Prover 1.5 achieves superior performance with a smaller compute budget. It solves \textbf{88\% of PutnamBench} (undergraduate-level), \textbf{80\% of Fate-H} (graduate-level), and \textbf{33\% of Fate-X} (PhD-level) problems. Notably, using our system, we solved \textbf{11 out of 12 problems} from Putnam 2025 within 9 hours. Our findings suggest that scaling learning from experience, driven by high-quality formal feedback, holds immense potential for the future of formal mathematical reasoning.
MolSculpt: Sculpting 3D Molecular Geometries from Chemical Syntax
Dec 09, 2025Abstract:Generating precise 3D molecular geometries is crucial for drug discovery and material science. While prior efforts leverage 1D representations like SELFIES to ensure molecular validity, they fail to fully exploit the rich chemical knowledge entangled within 1D models, leading to a disconnect between 1D syntactic generation and 3D geometric realization. To bridge this gap, we propose MolSculpt, a novel framework that "sculpts" 3D molecular geometries from chemical syntax. MolSculpt is built upon a frozen 1D molecular foundation model and a 3D molecular diffusion model. We introduce a set of learnable queries to extract inherent chemical knowledge from the foundation model, and a trainable projector then injects this cross-modal information into the conditioning space of the diffusion model to guide the 3D geometry generation. In this way, our model deeply integrates 1D latent chemical knowledge into the 3D generation process through end-to-end optimization. Experiments demonstrate that MolSculpt achieves state-of-the-art (SOTA) performance in \textit{de novo} 3D molecule generation and conditional 3D molecule generation, showing superior 3D fidelity and stability on both the GEOM-DRUGS and QM9 datasets. Code is available at https://github.com/SakuraTroyChen/MolSculpt.
A New Deep-learning-Based Approach For mRNA Optimization: High Fidelity, Computation Efficiency, and Multiple Optimization Factors
May 29, 2025Abstract:The mRNA optimization is critical for therapeutic and biotechnological applications, since sequence features directly govern protein expression levels and efficacy. However, current methods face significant challenges in simultaneously achieving three key objectives: (1) fidelity (preventing unintended amino acid changes), (2) computational efficiency (speed and scalability), and (3) the scope of optimization variables considered (multi-objective capability). Furthermore, existing methods often fall short of comprehensively incorporating the factors related to the mRNA lifecycle and translation process, including intrinsic mRNA sequence properties, secondary structure, translation elongation kinetics, and tRNA availability. To address these limitations, we introduce \textbf{RNop}, a novel deep learning-based method for mRNA optimization. We collect a large-scale dataset containing over 3 million sequences and design four specialized loss functions, the GPLoss, CAILoss, tAILoss, and MFELoss, which simultaneously enable explicit control over sequence fidelity while optimizing species-specific codon adaptation, tRNA availability, and desirable mRNA secondary structure features. Then, we demonstrate RNop's effectiveness through extensive in silico and in vivo experiments. RNop ensures high sequence fidelity, achieves significant computational throughput up to 47.32 sequences/s, and yields optimized mRNA sequences resulting in a significant increase in protein expression for functional proteins compared to controls. RNop surpasses current methodologies in both quantitative metrics and experimental validation, enlightening a new dawn for efficient and effective mRNA design. Code and models will be available at https://github.com/HudenJear/RPLoss.
AI-driven Prediction of Insulin Resistance in Normal Populations: Comparing Models and Criteria
Mar 07, 2025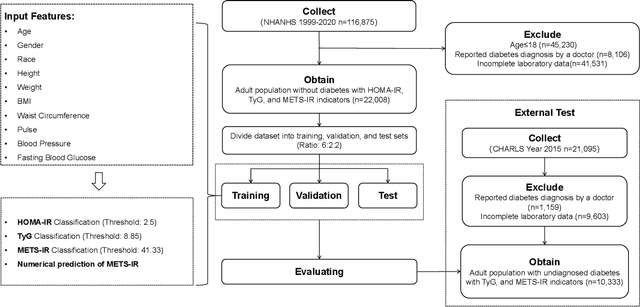
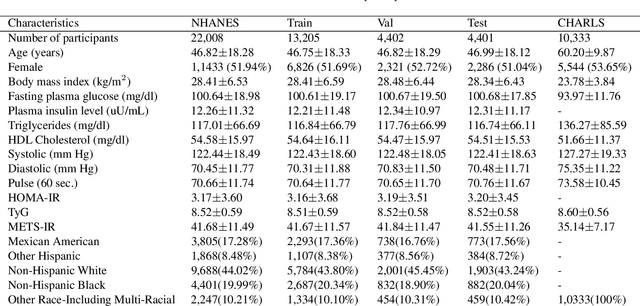
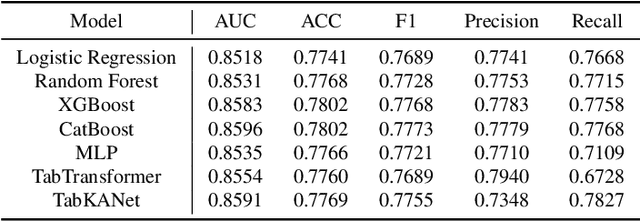
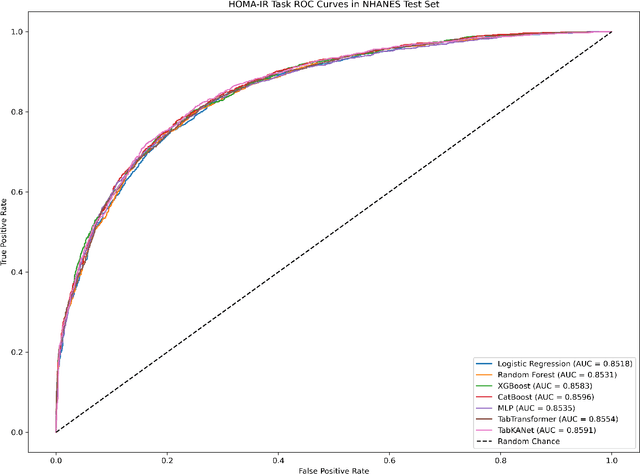
Abstract:Insulin resistance (IR) is a key precursor to diabetes and a significant risk factor for cardiovascular disease. Traditional IR assessment methods require multiple blood tests. We developed a simple AI model using only fasting blood glucose to predict IR in non-diabetic populations. Data from the NHANES (1999-2020) and CHARLS (2015) studies were used for model training and validation. Input features included age, gender, height, weight, blood pressure, waist circumference, and fasting blood glucose. The CatBoost algorithm achieved AUC values of 0.8596 (HOMA-IR) and 0.7777 (TyG index) in NHANES, with an external AUC of 0.7442 for TyG. For METS-IR prediction, the model achieved AUC values of 0.9731 (internal) and 0.9591 (external), with RMSE values of 3.2643 (internal) and 3.057 (external). SHAP analysis highlighted waist circumference as a key predictor of IR. This AI model offers a minimally invasive and effective tool for IR prediction, supporting early diabetes and cardiovascular disease prevention.
Acquire Precise and Comparable Fundus Image Quality Score: FTHNet and FQS Dataset
Nov 19, 2024Abstract:The retinal fundus images are utilized extensively in the diagnosis, and their quality can directly affect the diagnosis results. However, due to the insufficient dataset and algorithm application, current fundus image quality assessment (FIQA) methods are not powerful enough to meet ophthalmologists` demands. In this paper, we address the limitations of datasets and algorithms in FIQA. First, we establish a new FIQA dataset, Fundus Quality Score(FQS), which includes 2246 fundus images with two labels: a continuous Mean Opinion Score varying from 0 to 100 and a three-level quality label. Then, we propose a FIQA Transformer-based Hypernetwork (FTHNet) to solve these tasks with regression results rather than classification results in conventional FIQA works. The FTHNet is optimized for the FIQA tasks with extensive experiments. Results on our FQS dataset show that the FTHNet can give quality scores for fundus images with PLCC of 0.9423 and SRCC of 0.9488, significantly outperforming other methods with fewer parameters and less computation complexity.We successfully build a dataset and model addressing the problems of current FIQA methods. Furthermore, the model deployment experiments demonstrate its potential in automatic medical image quality control. All experiments are carried out with 10-fold cross-validation to ensure the significance of the results.
Versatile Cataract Fundus Image Restoration Model Utilizing Unpaired Cataract and High-quality Images
Nov 19, 2024



Abstract:Cataract is one of the most common blinding eye diseases and can be treated by surgery. However, because cataract patients may also suffer from other blinding eye diseases, ophthalmologists must diagnose them before surgery. The cloudy lens of cataract patients forms a hazy degeneration in the fundus images, making it challenging to observe the patient's fundus vessels, which brings difficulties to the diagnosis process. To address this issue, this paper establishes a new cataract image restoration method named Catintell. It contains a cataract image synthesizing model, Catintell-Syn, and a restoration model, Catintell-Res. Catintell-Syn uses GAN architecture with fully unsupervised data to generate paired cataract-like images with realistic style and texture rather than the conventional Gaussian degradation algorithm. Meanwhile, Catintell-Res is an image restoration network that can improve the quality of real cataract fundus images using the knowledge learned from synthetic cataract images. Extensive experiments show that Catintell-Res outperforms other cataract image restoration methods in PSNR with 39.03 and SSIM with 0.9476. Furthermore, the universal restoration ability that Catintell-Res gained from unpaired cataract images can process cataract images from various datasets. We hope the models can help ophthalmologists identify other blinding eye diseases of cataract patients and inspire more medical image restoration methods in the future.
TabKANet: Tabular Data Modelling with Kolmogorov-Arnold Network and Transformer
Sep 13, 2024



Abstract:Tabular data is the most common type of data in real-life scenarios. In this study, we propose a method based on the TabKANet architecture, which utilizes the Kolmogorov-Arnold network to encode numerical features and merge them with categorical features, enabling unified modeling of tabular data on the Transformer architecture. This model demonstrates outstanding performance in six widely used binary classification tasks, suggesting that TabKANet has the potential to become a standard approach for tabular modeling, surpassing traditional neural networks. Furthermore, this research reveals the significant advantages of the Kolmogorov-Arnold network in encoding numerical features. The code of our work is available at https://github.com/tsinghuamedgao20/TabKANet.
Crystals with Transformers on Graphs, for Prediction of Unconventional Crystal Material Properties and the Benchmark
Jul 23, 2024Abstract:The ionic bonding across the lattice and ordered microscopic structures endow crystals with unique symmetry and determine their macroscopic properties. Unconventional crystals, in particular, exhibit non-traditional lattice structures or possess exotic physical properties, making them intriguing subjects for investigation. Therefore, to accurately predict the physical and chemical properties of crystals, it is crucial to consider long-range orders. While GNN excels at capturing the local environment of atoms in crystals, they often face challenges in effectively capturing longer-ranged interactions due to their limited depth. In this paper, we propose CrysToGraph ($\textbf{Crys}$tals with $\textbf{T}$ransformers $\textbf{o}$n $\textbf{Graph}$s), a novel transformer-based geometric graph network designed specifically for unconventional crystalline systems, and UnconvBench, a comprehensive benchmark to evaluate models' predictive performance on unconventional crystal materials such as defected crystals, low-dimension crystals and MOF. CrysToGraph effectively captures short-range interactions with transformer-based graph convolution blocks as well as long-range interactions with graph-wise transformer blocks. CrysToGraph proofs its effectiveness in modelling unconventional crystal materials in multiple tasks, and moreover, it outperforms most existing methods, achieving new state-of-the-art results on the benchmarks of both unconventional crystals and traditional crystals.
BAMBOO: a predictive and transferable machine learning force field framework for liquid electrolyte development
Apr 12, 2024Abstract:Despite the widespread applications of machine learning force field (MLFF) on solids and small molecules, there is a notable gap in applying MLFF to complex liquid electrolytes. In this work, we introduce BAMBOO (ByteDance AI Molecular Simulation Booster), a novel framework for molecular dynamics (MD) simulations, with a demonstration of its capabilities in the context of liquid electrolytes for lithium batteries. We design a physics-inspired graph equivariant transformer architecture as the backbone of BAMBOO to learn from quantum mechanical simulations. Additionally, we pioneer an ensemble knowledge distillation approach and apply it on MLFFs to improve the stability of MD simulations. Finally, we propose the density alignment algorithm to align BAMBOO with experimental measurements. BAMBOO demonstrates state-of-the-art accuracy in predicting key electrolyte properties such as density, viscosity, and ionic conductivity across various solvents and salt combinations. Our current model, trained on more than 15 chemical species, achieves the average density error of 0.01 g/cm$^3$ on various compositions compared with experimental data. Moreover, our model demonstrates transferability to molecules not included in the quantum mechanical dataset. We envision this work as paving the way to a "universal MLFF" capable of simulating properties of common organic liquids.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge