Qianjin Feng
Cross-modal ultra-scale learning with tri-modalities of renal biopsy images for glomerular multi-disease auxiliary diagnosis
Dec 17, 2025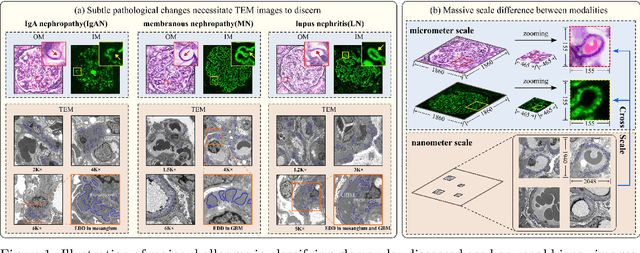
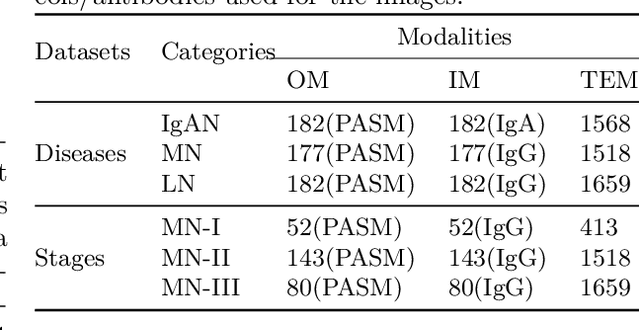
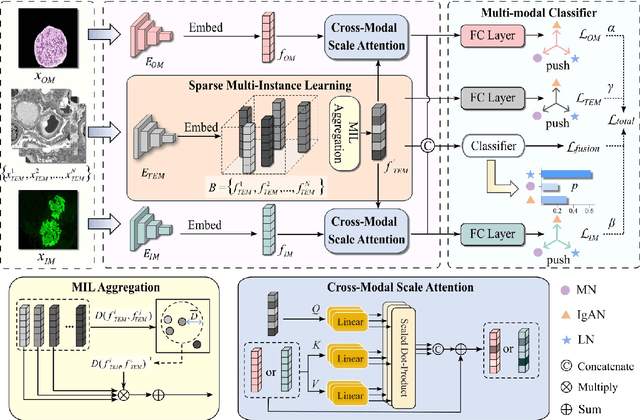
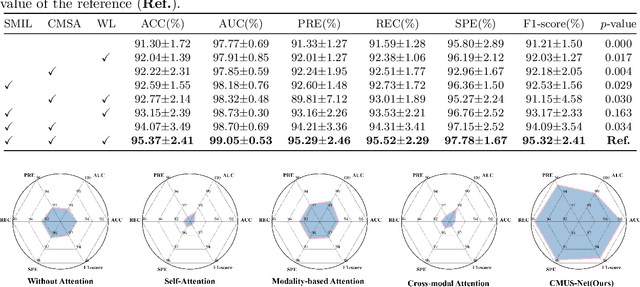
Abstract:Constructing a multi-modal automatic classification model based on three types of renal biopsy images can assist pathologists in glomerular multi-disease identification. However, the substantial scale difference between transmission electron microscopy (TEM) image features at the nanoscale and optical microscopy (OM) or immunofluorescence microscopy (IM) images at the microscale poses a challenge for existing multi-modal and multi-scale models in achieving effective feature fusion and improving classification accuracy. To address this issue, we propose a cross-modal ultra-scale learning network (CMUS-Net) for the auxiliary diagnosis of multiple glomerular diseases. CMUS-Net utilizes multiple ultrastructural information to bridge the scale difference between nanometer and micrometer images. Specifically, we introduce a sparse multi-instance learning module to aggregate features from TEM images. Furthermore, we design a cross-modal scale attention module to facilitate feature interaction, enhancing pathological semantic information. Finally, multiple loss functions are combined, allowing the model to weigh the importance among different modalities and achieve precise classification of glomerular diseases. Our method follows the conventional process of renal biopsy pathology diagnosis and, for the first time, performs automatic classification of multiple glomerular diseases including IgA nephropathy (IgAN), membranous nephropathy (MN), and lupus nephritis (LN) based on images from three modalities and two scales. On an in-house dataset, CMUS-Net achieves an ACC of 95.37+/-2.41%, an AUC of 99.05+/-0.53%, and an F1-score of 95.32+/-2.41%. Extensive experiments demonstrate that CMUS-Net outperforms other well-known multi-modal or multi-scale methods and show its generalization capability in staging MN. Code is available at https://github.com/SMU-GL-Group/MultiModal_lkx/tree/main.
DCMIL: A Progressive Representation Learning Model of Whole Slide Images for Cancer Prognosis Analysis
Oct 16, 2025Abstract:The burgeoning discipline of computational pathology shows promise in harnessing whole slide images (WSIs) to quantify morphological heterogeneity and develop objective prognostic modes for human cancers. However, progress is impeded by the computational bottleneck of gigapixel-size inputs and the scarcity of dense manual annotations. Current methods often overlook fine-grained information across multi-magnification WSIs and variations in tumor microenvironments. Here, we propose an easy-to-hard progressive representation learning model, termed dual-curriculum contrastive multi-instance learning (DCMIL), to efficiently process WSIs for cancer prognosis. The model does not rely on dense annotations and enables the direct transformation of gigapixel-size WSIs into outcome predictions. Extensive experiments on twelve cancer types (5,954 patients, 12.54 million tiles) demonstrate that DCMIL outperforms standard WSI-based prognostic models. Additionally, DCMIL identifies fine-grained prognosis-salient regions, provides robust instance uncertainty estimation, and captures morphological differences between normal and tumor tissues, with the potential to generate new biological insights. All codes have been made publicly accessible at https://github.com/tuuuc/DCMIL.
SpineMamba: Enhancing 3D Spinal Segmentation in Clinical Imaging through Residual Visual Mamba Layers and Shape Priors
Aug 28, 2024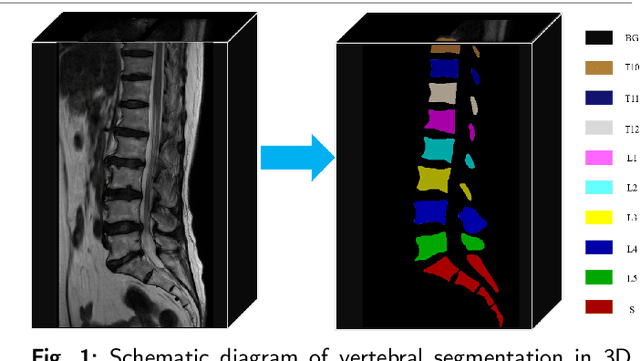
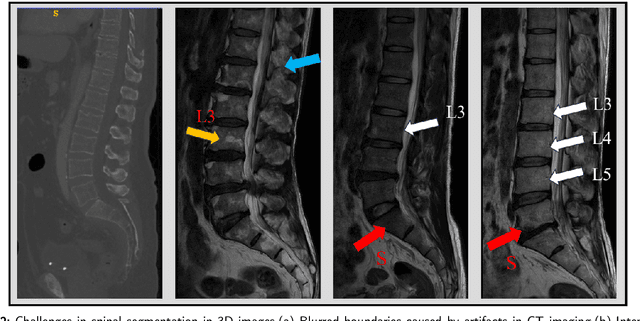
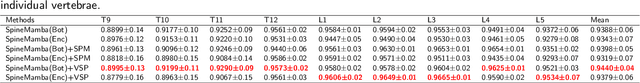
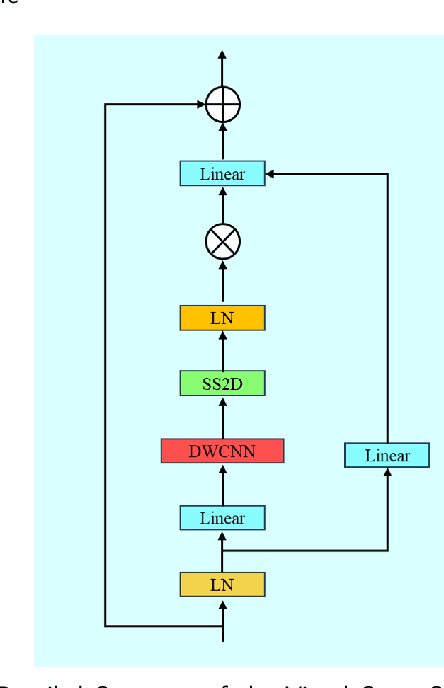
Abstract:Accurate segmentation of 3D clinical medical images is critical in the diagnosis and treatment of spinal diseases. However, the inherent complexity of spinal anatomy and uncertainty inherent in current imaging technologies, poses significant challenges for semantic segmentation of spinal images. Although convolutional neural networks (CNNs) and Transformer-based models have made some progress in spinal segmentation, their limitations in handling long-range dependencies hinder further improvements in segmentation accuracy.To address these challenges, we introduce a residual visual Mamba layer to effectively capture and model the deep semantic features and long-range spatial dependencies of 3D spinal data. To further enhance the structural semantic understanding of the vertebrae, we also propose a novel spinal shape prior module that captures specific anatomical information of the spine from medical images, significantly enhancing the model's ability to extract structural semantic information of the vertebrae. Comparative and ablation experiments on two datasets demonstrate that SpineMamba outperforms existing state-of-the-art models. On the CT dataset, the average Dice similarity coefficient for segmentation reaches as high as 94.40, while on the MR dataset, it reaches 86.95. Notably, compared to the renowned nnU-Net, SpineMamba achieves superior segmentation performance, exceeding it by up to 2 percentage points. This underscores its accuracy, robustness, and excellent generalization capabilities.
Performance of Medical Image Fusion in High-level Analysis Tasks: A Mutual Enhancement Framework for Unaligned PAT and MRI Image Fusion
Jul 04, 2024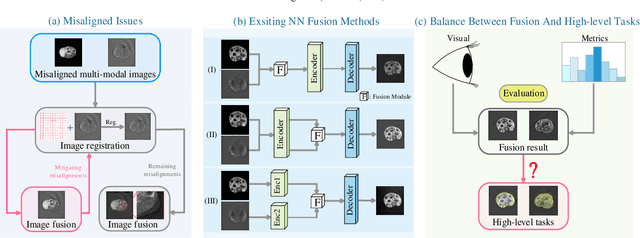
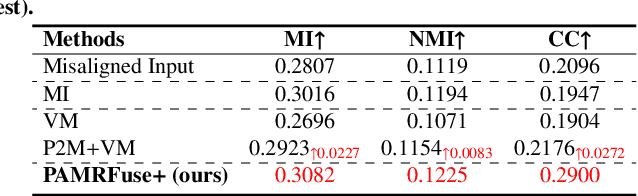
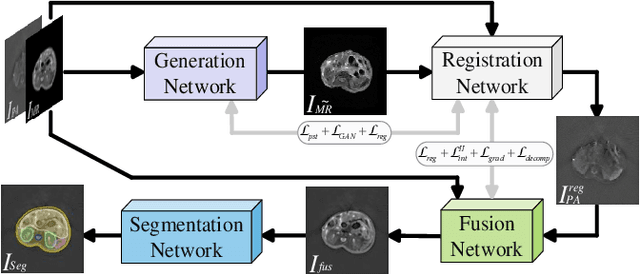
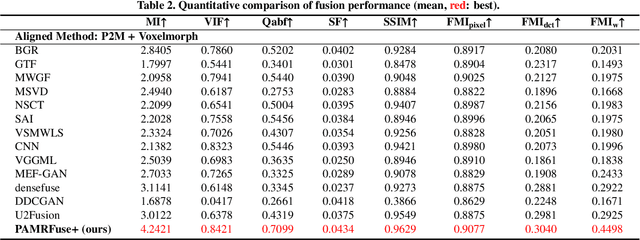
Abstract:Photoacoustic tomography (PAT) offers optical contrast, whereas magnetic resonance imaging (MRI) excels in imaging soft tissue and organ anatomy. The fusion of PAT with MRI holds promising application prospects due to their complementary advantages. Existing image fusion have made considerable progress in pre-registered images, yet spatial deformations are difficult to avoid in medical imaging scenarios. More importantly, current algorithms focus on visual quality and statistical metrics, thus overlooking the requirements of high-level tasks. To address these challenges, we proposes a unsupervised fusion model, termed PAMRFuse+, which integrates image generation and registration. Specifically, a cross-modal style transfer network is introduced to simplify cross-modal registration to single-modal registration. Subsequently, a multi-level registration network is employed to predict displacement vector fields. Furthermore, a dual-branch feature decomposition fusion network is proposed to address the challenges of cross-modal feature modeling and decomposition by integrating modality-specific and modality-shared features. PAMRFuse+ achieves satisfactory results in registering and fusing unaligned PAT-MRI datasets. Moreover, for the first time, we evaluate the performance of medical image fusion with contour segmentation and multi-organ instance segmentation. Extensive experimental demonstrations reveal the advantages of PAMRFuse+ in improving the performance of medical image analysis tasks.
A multi-stage semi-supervised learning for ankle fracture classification on CT images
Mar 29, 2024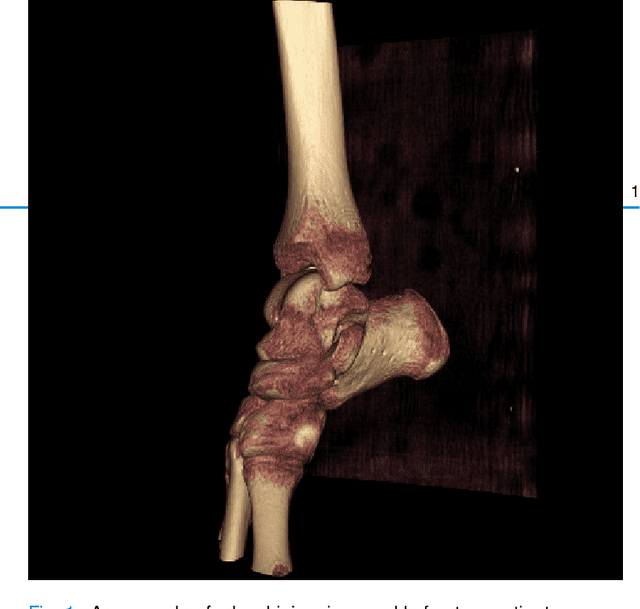
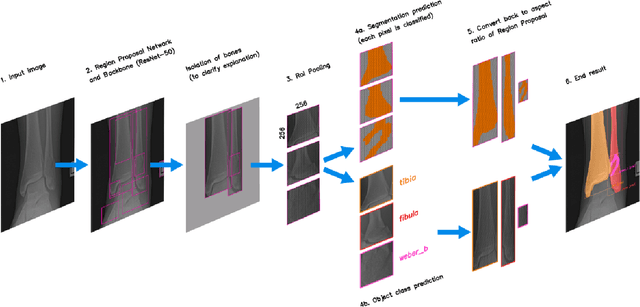
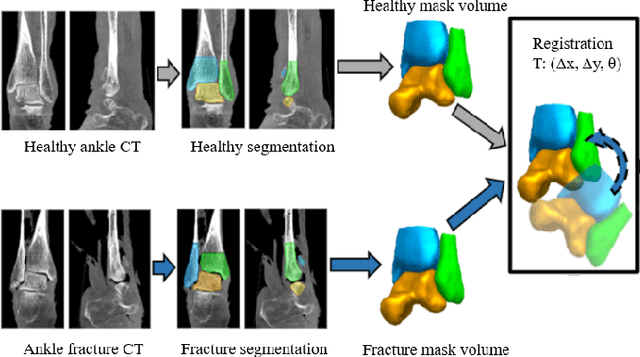
Abstract:Because of the complicated mechanism of ankle injury, it is very difficult to diagnose ankle fracture in clinic. In order to simplify the process of fracture diagnosis, an automatic diagnosis model of ankle fracture was proposed. Firstly, a tibia-fibula segmentation network is proposed for the joint tibiofibular region of the ankle joint, and the corresponding segmentation dataset is established on the basis of fracture data. Secondly, the image registration method is used to register the bone segmentation mask with the normal bone mask. Finally, a semi-supervised classifier is constructed to make full use of a large number of unlabeled data to classify ankle fractures. Experiments show that the proposed method can segment fractures with fracture lines accurately and has better performance than the general method. At the same time, this method is superior to classification network in several indexes.
Decomposing and Coupling Saliency Map for Lesion Segmentation in Ultrasound Images
Aug 02, 2023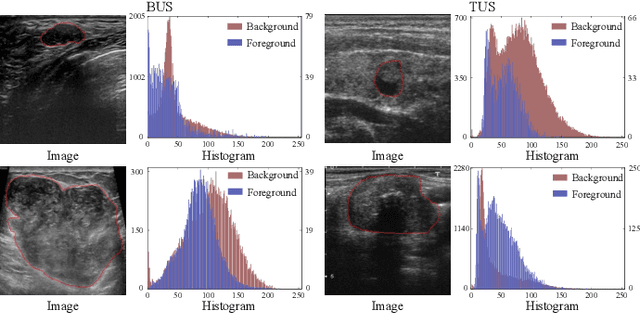

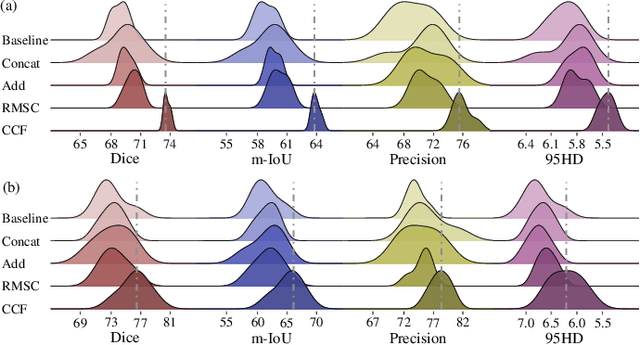
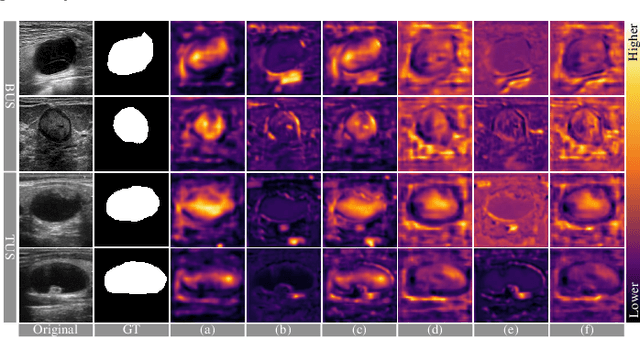
Abstract:Complex scenario of ultrasound image, in which adjacent tissues (i.e., background) share similar intensity with and even contain richer texture patterns than lesion region (i.e., foreground), brings a unique challenge for accurate lesion segmentation. This work presents a decomposition-coupling network, called DC-Net, to deal with this challenge in a (foreground-background) saliency map disentanglement-fusion manner. The DC-Net consists of decomposition and coupling subnets, and the former preliminarily disentangles original image into foreground and background saliency maps, followed by the latter for accurate segmentation under the assistance of saliency prior fusion. The coupling subnet involves three aspects of fusion strategies, including: 1) regional feature aggregation (via differentiable context pooling operator in the encoder) to adaptively preserve local contextual details with the larger receptive field during dimension reduction; 2) relation-aware representation fusion (via cross-correlation fusion module in the decoder) to efficiently fuse low-level visual characteristics and high-level semantic features during resolution restoration; 3) dependency-aware prior incorporation (via coupler) to reinforce foreground-salient representation with the complementary information derived from background representation. Furthermore, a harmonic loss function is introduced to encourage the network to focus more attention on low-confidence and hard samples. The proposed method is evaluated on two ultrasound lesion segmentation tasks, which demonstrates the remarkable performance improvement over existing state-of-the-art methods.
Multi-View Imputation and Cross-Attention Network Based on Incomplete Longitudinal and Multi-Modal Data for Alzheimer's Disease Prediction
Jun 16, 2022



Abstract:Longitudinal variations and complementary information inherent in longitudinal and multi-modal data play an important role in Alzheimer's disease (AD) prediction, particularly in identifying subjects with mild cognitive impairment who are about to have AD. However, longitudinal and multi-modal data may have missing data, which hinders the effective application of these data. Additionally, previous longitudinal studies require existing longitudinal data to achieve prediction, but AD prediction is expected to be conducted at patients' baseline visit (BL) in clinical practice. Thus, we proposed a multi-view imputation and cross-attention network (MCNet) to integrate data imputation and AD prediction in a unified framework and achieve accurate AD prediction. First, a multi-view imputation method combined with adversarial learning, which can handle a wide range of missing data situations and reduce imputation errors, was presented. Second, two cross-attention blocks were introduced to exploit the potential associations in longitudinal and multi-modal data. Finally, a multi-task learning model was built for data imputation, longitudinal classification, and AD prediction tasks. When the model was properly trained, the disease progression information learned from longitudinal data can be leveraged by BL data to improve AD prediction. The proposed method was tested on two independent testing sets and single-model data at BL to verify its effectiveness and flexibility on AD prediction. Results showed that MCNet outperformed several state-of-the-art methods. Moreover, the interpretability of MCNet was presented. Thus, our MCNet is a tool with a great application potential in longitudinal and multi-modal data analysis for AD prediction. Codes are available at https://github.com/Meiyan88/MCNET.
Semi-Supervised Hybrid Spine Network for Segmentation of Spine MR Images
Mar 23, 2022



Abstract:Automatic segmentation of vertebral bodies (VBs) and intervertebral discs (IVDs) in 3D magnetic resonance (MR) images is vital in diagnosing and treating spinal diseases. However, segmenting the VBs and IVDs simultaneously is not trivial. Moreover, problems exist, including blurry segmentation caused by anisotropy resolution, high computational cost, inter-class similarity and intra-class variability, and data imbalances. We proposed a two-stage algorithm, named semi-supervised hybrid spine network (SSHSNet), to address these problems by achieving accurate simultaneous VB and IVD segmentation. In the first stage, we constructed a 2D semi-supervised DeepLabv3+ by using cross pseudo supervision to obtain intra-slice features and coarse segmentation. In the second stage, a 3D full-resolution patch-based DeepLabv3+ was built. This model can be used to extract inter-slice information and combine the coarse segmentation and intra-slice features provided from the first stage. Moreover, a cross tri-attention module was applied to compensate for the loss of inter-slice and intra-slice information separately generated from 2D and 3D networks, thereby improving feature representation ability and achieving satisfactory segmentation results. The proposed SSHSNet was validated on a publicly available spine MR image dataset, and remarkable segmentation performance was achieved. Moreover, results show that the proposed method has great potential in dealing with the data imbalance problem. Based on previous reports, few studies have incorporated a semi-supervised learning strategy with a cross attention mechanism for spine segmentation. Therefore, the proposed method may provide a useful tool for spine segmentation and aid clinically in spinal disease diagnoses and treatments. Codes are publicly available at: https://github.com/Meiyan88/SSHSNet.
Unsupervised Deformable Medical Image Registration via Pyramidal Residual Deformation Fields Estimation
Apr 16, 2020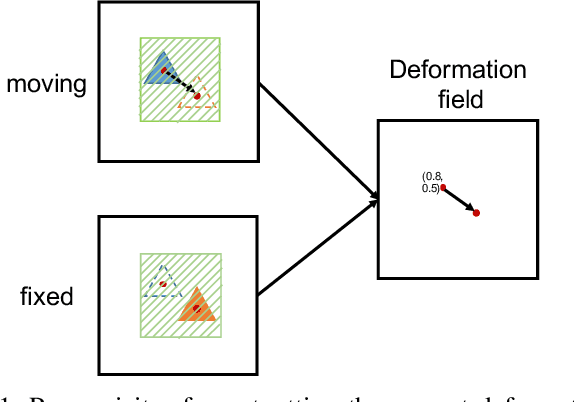
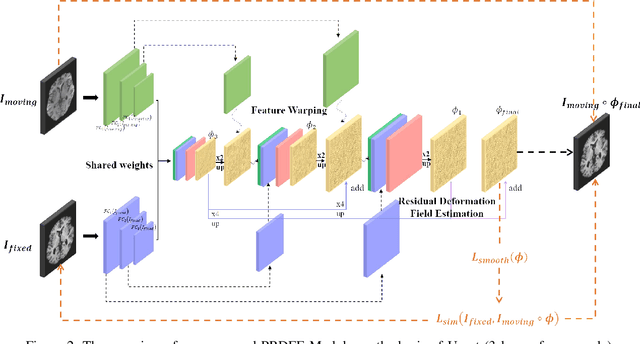
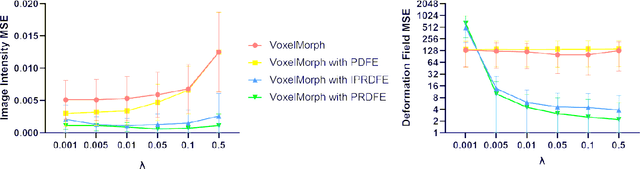
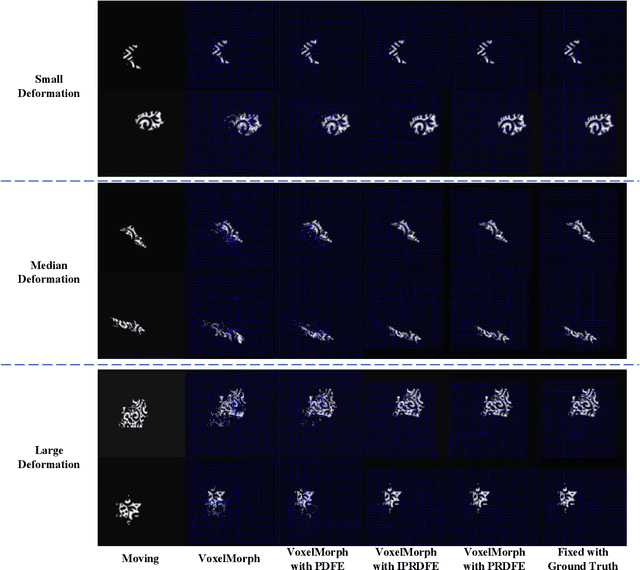
Abstract:Deformation field estimation is an important and challenging issue in many medical image registration applications. In recent years, deep learning technique has become a promising approach for simplifying registration problems, and has been gradually applied to medical image registration. However, most existing deep learning registrations do not consider the problem that when the receptive field cannot cover the corresponding features in the moving image and the fixed image, it cannot output accurate displacement values. In fact, due to the limitation of the receptive field, the 3 x 3 kernel has difficulty in covering the corresponding features at high/original resolution. Multi-resolution and multi-convolution techniques can improve but fail to avoid this problem. In this study, we constructed pyramidal feature sets on moving and fixed images and used the warped moving and fixed features to estimate their "residual" deformation field at each scale, called the Pyramidal Residual Deformation Field Estimation module (PRDFE-Module). The "total" deformation field at each scale was computed by upsampling and weighted summing all the "residual" deformation fields at all its previous scales, which can effectively and accurately transfer the deformation fields from low resolution to high resolution and is used for warping the moving features at each scale. Simulation and real brain data results show that our method improves the accuracy of the registration and the rationality of the deformation field.
CF2-Net: Coarse-to-Fine Fusion Convolutional Network for Breast Ultrasound Image Segmentation
Mar 23, 2020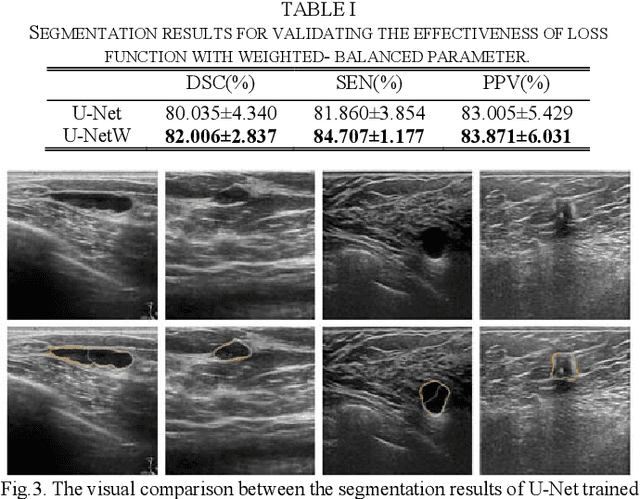
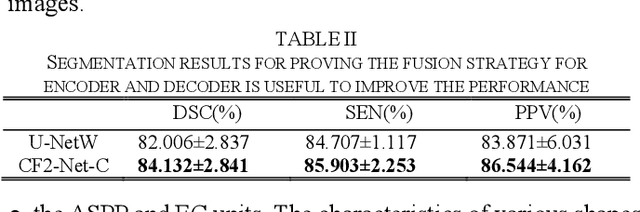
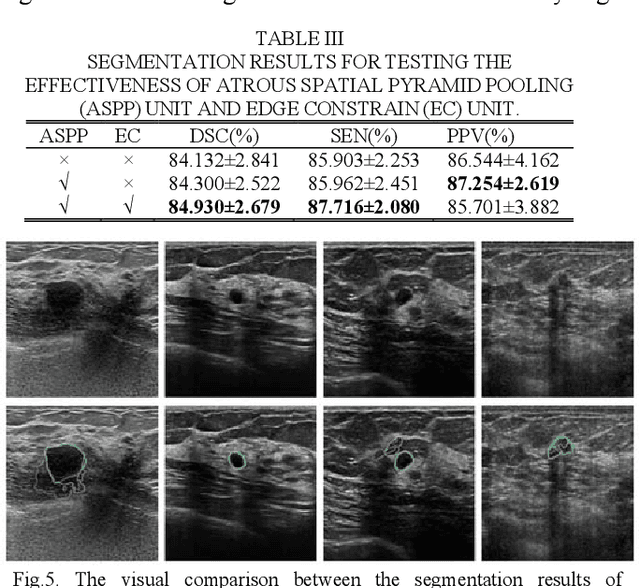
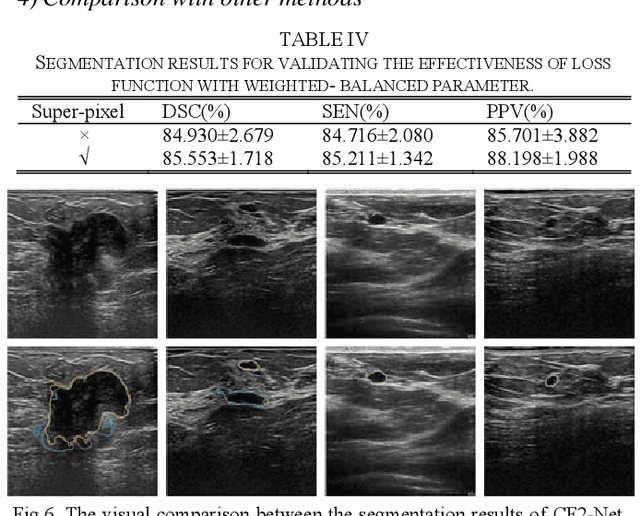
Abstract:Breast ultrasound (BUS) image segmentation plays a crucial role in a computer-aided diagnosis system, which is regarded as a useful tool to help increase the accuracy of breast cancer diagnosis. Recently, many deep learning methods have been developed for segmentation of BUS image and show some advantages compared with conventional region-, model-, and traditional learning-based methods. However, previous deep learning methods typically use skip-connection to concatenate the encoder and decoder, which might not make full fusion of coarse-to-fine features from encoder and decoder. Since the structure and edge of lesion in BUS image are common blurred, these would make it difficult to learn the discriminant information of structure and edge, and reduce the performance. To this end, we propose and evaluate a coarse-to-fine fusion convolutional network (CF2-Net) based on a novel feature integration strategy (forming an 'E'-like type) for BUS image segmentation. To enhance contour and provide structural information, we concatenate a super-pixel image and the original image as the input of CF2-Net. Meanwhile, to highlight the differences in the lesion regions with variable sizes and relieve the imbalance issue, we further design a weighted-balanced loss function to train the CF2-Net effectively. The proposed CF2-Net was evaluated on an open dataset by using four-fold cross validation. The results of the experiment demonstrate that the CF2-Net obtains state-of-the-art performance when compared with other deep learning-based methods
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge