Haoran Lai
Med3D-R1: Incentivizing Clinical Reasoning in 3D Medical Vision-Language Models for Abnormality Diagnosis
Feb 01, 2026Abstract:Developing 3D vision-language models with robust clinical reasoning remains a challenge due to the inherent complexity of volumetric medical imaging, the tendency of models to overfit superficial report patterns, and the lack of interpretability-aware reward designs. In this paper, we propose Med3D-R1, a reinforcement learning framework with a two-stage training process: Supervised Fine-Tuning (SFT) and Reinforcement Learning (RL). During SFT stage, we introduce a residual alignment mechanism to bridge the gap between high-dimensional 3D features and textual embeddings, and an abnormality re-weighting strategy to emphasize clinically informative tokens and reduce structural bias in reports. In RL stage, we redesign the consistency reward to explicitly promote coherent, step-by-step diagnostic reasoning. We evaluate our method on medical multiple-choice visual question answering using two 3D diagnostic benchmarks, CT-RATE and RAD-ChestCT, where our model attains state-of-the-art accuracies of 41.92\% on CT-RATE and 44.99\% on RAD-ChestCT. These results indicate improved abnormality diagnosis and clinical reasoning and outperform prior methods on both benchmarks. Overall, our approach holds promise for enhancing real-world diagnostic workflows by enabling more reliable and transparent 3D medical vision-language systems.
More performant and scalable: Rethinking contrastive vision-language pre-training of radiology in the LLM era
Sep 16, 2025Abstract:The emergence of Large Language Models (LLMs) presents unprecedented opportunities to revolutionize medical contrastive vision-language pre-training. In this paper, we show how LLMs can facilitate large-scale supervised pre-training, thereby advancing vision-language alignment. We begin by demonstrate that modern LLMs can automatically extract diagnostic labels from radiology reports with remarkable precision (>96\% AUC in our experiments) without complex prompt engineering, enabling the creation of large-scale "silver-standard" datasets at a minimal cost (~\$3 for 50k CT image-report pairs). Further, we find that vision encoder trained on this "silver-standard" dataset achieves performance comparable to those trained on labels extracted by specialized BERT-based models, thereby democratizing the access to large-scale supervised pre-training. Building on this foundation, we proceed to reveal that supervised pre-training fundamentally improves contrastive vision-language alignment. Our approach achieves state-of-the-art performance using only a 3D ResNet-18 with vanilla CLIP training, including 83.8\% AUC for zero-shot diagnosis on CT-RATE, 77.3\% AUC on RAD-ChestCT, and substantial improvements in cross-modal retrieval (MAP@50=53.7\% for image-image, Recall@100=52.2\% for report-image). These results demonstrate the potential of utilizing LLMs to facilitate {\bf more performant and scalable} medical AI systems. Our code is avaiable at https://github.com/SadVoxel/More-performant-and-scalable.
SimCroP: Radiograph Representation Learning with Similarity-driven Cross-granularity Pre-training
Sep 10, 2025Abstract:Medical vision-language pre-training shows great potential in learning representative features from massive paired radiographs and reports. However, in computed tomography (CT) scans, the distribution of lesions which contain intricate structures is characterized by spatial sparsity. Besides, the complex and implicit relationships between different pathological descriptions in each sentence of the report and their corresponding sub-regions in radiographs pose additional challenges. In this paper, we propose a Similarity-Driven Cross-Granularity Pre-training (SimCroP) framework on chest CTs, which combines similarity-driven alignment and cross-granularity fusion to improve radiograph interpretation. We first leverage multi-modal masked modeling to optimize the encoder for understanding precise low-level semantics from radiographs. Then, similarity-driven alignment is designed to pre-train the encoder to adaptively select and align the correct patches corresponding to each sentence in reports. The cross-granularity fusion module integrates multimodal information across instance level and word-patch level, which helps the model better capture key pathology structures in sparse radiographs, resulting in improved performance for multi-scale downstream tasks. SimCroP is pre-trained on a large-scale paired CT-reports dataset and validated on image classification and segmentation tasks across five public datasets. Experimental results demonstrate that SimCroP outperforms both cutting-edge medical self-supervised learning methods and medical vision-language pre-training methods. Codes and models are available at https://github.com/ToniChopp/SimCroP.
Bridged Semantic Alignment for Zero-shot 3D Medical Image Diagnosis
Jan 07, 2025Abstract:3D medical images such as Computed tomography (CT) are widely used in clinical practice, offering a great potential for automatic diagnosis. Supervised learning-based approaches have achieved significant progress but rely heavily on extensive manual annotations, limited by the availability of training data and the diversity of abnormality types. Vision-language alignment (VLA) offers a promising alternative by enabling zero-shot learning without additional annotations. However, we empirically discover that the visual and textural embeddings after alignment endeavors from existing VLA methods form two well-separated clusters, presenting a wide gap to be bridged. To bridge this gap, we propose a Bridged Semantic Alignment (BrgSA) framework. First, we utilize a large language model to perform semantic summarization of reports, extracting high-level semantic information. Second, we design a Cross-Modal Knowledge Interaction (CMKI) module that leverages a cross-modal knowledge bank as a semantic bridge, facilitating interaction between the two modalities, narrowing the gap, and improving their alignment. To comprehensively evaluate our method, we construct a benchmark dataset that includes 15 underrepresented abnormalities as well as utilize two existing benchmark datasets. Experimental results demonstrate that BrgSA achieves state-of-the-art performances on both public benchmark datasets and our custom-labeled dataset, with significant improvements in zero-shot diagnosis of underrepresented abnormalities.
E3D-GPT: Enhanced 3D Visual Foundation for Medical Vision-Language Model
Oct 18, 2024
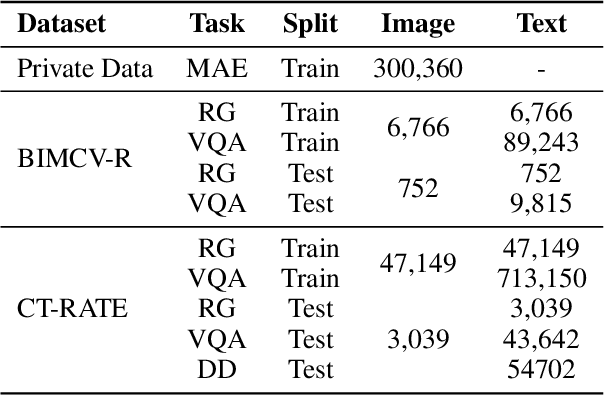
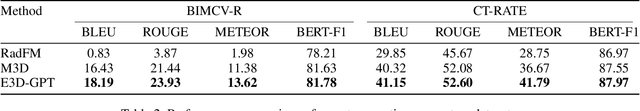

Abstract:The development of 3D medical vision-language models holds significant potential for disease diagnosis and patient treatment. However, compared to 2D medical images, 3D medical images, such as CT scans, face challenges related to limited training data and high dimension, which severely restrict the progress of 3D medical vision-language models. To address these issues, we collect a large amount of unlabeled 3D CT data and utilize self-supervised learning to construct a 3D visual foundation model for extracting 3D visual features. Then, we apply 3D spatial convolutions to aggregate and project high-level image features, reducing computational complexity while preserving spatial information. We also construct two instruction-tuning datasets based on BIMCV-R and CT-RATE to fine-tune the 3D vision-language model. Our model demonstrates superior performance compared to existing methods in report generation, visual question answering, and disease diagnosis. Code and data will be made publicly available soon.
CARZero: Cross-Attention Alignment for Radiology Zero-Shot Classification
Feb 27, 2024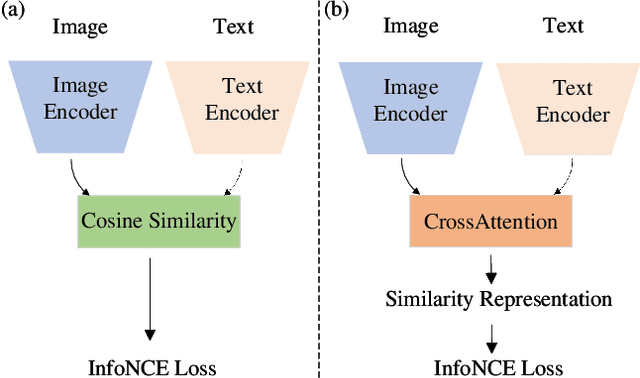
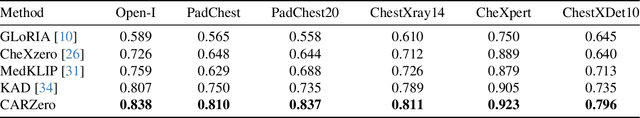
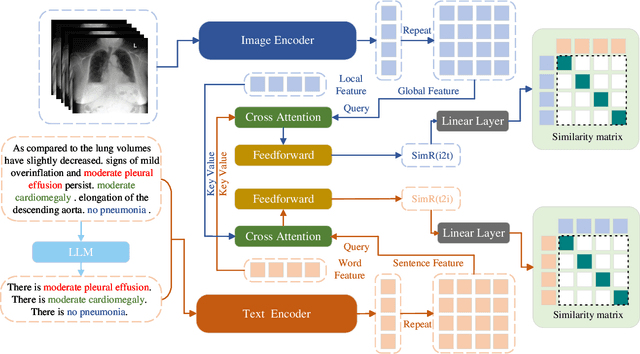
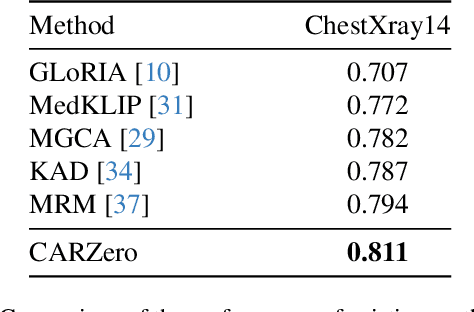
Abstract:The advancement of Zero-Shot Learning in the medical domain has been driven forward by using pre-trained models on large-scale image-text pairs, focusing on image-text alignment. However, existing methods primarily rely on cosine similarity for alignment, which may not fully capture the complex relationship between medical images and reports. To address this gap, we introduce a novel approach called Cross-Attention Alignment for Radiology Zero-Shot Classification (CARZero). Our approach innovatively leverages cross-attention mechanisms to process image and report features, creating a Similarity Representation that more accurately reflects the intricate relationships in medical semantics. This representation is then linearly projected to form an image-text similarity matrix for cross-modality alignment. Additionally, recognizing the pivotal role of prompt selection in zero-shot learning, CARZero incorporates a Large Language Model-based prompt alignment strategy. This strategy standardizes diverse diagnostic expressions into a unified format for both training and inference phases, overcoming the challenges of manual prompt design. Our approach is simple yet effective, demonstrating state-of-the-art performance in zero-shot classification on five official chest radiograph diagnostic test sets, including remarkable results on datasets with long-tail distributions of rare diseases. This achievement is attributed to our new image-text alignment strategy, which effectively addresses the complex relationship between medical images and reports.
ECAMP: Entity-centered Context-aware Medical Vision Language Pre-training
Dec 20, 2023Abstract:Despite significant advancements in medical vision-language pre-training, existing methods have largely overlooked the inherent entity-specific context within radiology reports and the complex cross-modality contextual relationships between text and images. To close this gap, we propose a novel Entity-centered Context-aware Medical Vision-language Pre-training (ECAMP) framework, which is designed to enable a more entity-centered and context-sensitive interpretation of medical data. Utilizing the recent powerful large language model, we distill entity-centered context from medical reports, which enables ECAMP to gain more effective supervision from the text modality. By further pre-training our model with carefully designed entity-aware, context-enhanced masked language modeling and context-guided super-resolution tasks, ECAMP significantly refines the interplay between text and image modalities, leading to an enhanced ability to extract entity-centered contextual features. Besides, our proposed multi-scale context fusion design also improves the semantic integration of both coarse and fine-level image representations, prompting better performance for multi-scale downstream applications. Combining these components leads to significant performance leaps over current state-of-the-art methods and establishes a new standard for cross-modality learning in medical imaging, whose effectiveness is demonstrated by our extensive experiments on various tasks including classification, segmentation, and detection across several public datasets. Code and models are available at https://github.com/ToniChopp/ECAMP.
Long-tailed multi-label classification with noisy label of thoracic diseases from chest X-ray
Nov 29, 2023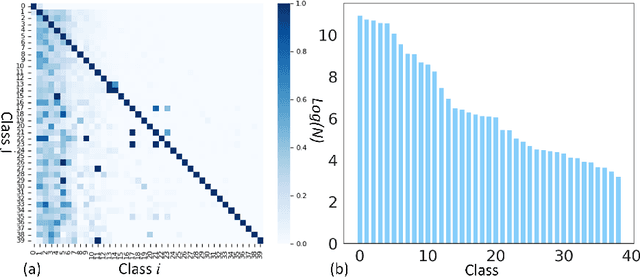
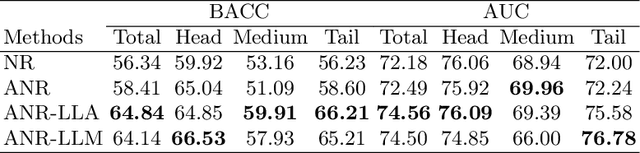
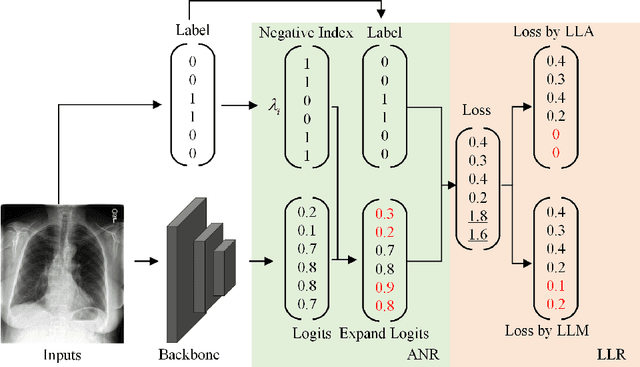
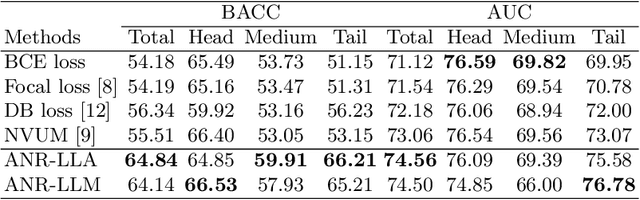
Abstract:Chest X-rays (CXR) often reveal rare diseases, demanding precise diagnosis. However, current computer-aided diagnosis (CAD) methods focus on common diseases, leading to inadequate detection of rare conditions due to the absence of comprehensive datasets. To overcome this, we present a novel benchmark for long-tailed multi-label classification in CXRs, encapsulating both common and rare thoracic diseases. Our approach includes developing the "LTML-MIMIC-CXR" dataset, an augmentation of MIMIC-CXR with 26 additional rare diseases. We propose a baseline method for this classification challenge, integrating adaptive negative regularization to address negative logits' over-suppression in tail classes, and a large loss reconsideration strategy for correcting noisy labels from automated annotations. Our evaluation on LTML-MIMIC-CXR demonstrates significant advancements in rare disease detection. This work establishes a foundation for robust CAD methods, achieving a balance in identifying a spectrum of thoracic diseases in CXRs. Access to our code and dataset is provided at:https://github.com/laihaoran/LTML-MIMIC-CXR.
DLUNet: Semi-supervised Learning based Dual-Light UNet for Multi-organ Segmentation
Sep 22, 2022



Abstract:The manual ground truth of abdominal multi-organ is labor-intensive. In order to make full use of CT data, we developed a semi-supervised learning based dual-light UNet. In the training phase, it consists of two light UNets, which make full use of label and unlabeled data simultaneously by using consistent-based learning. Moreover, separable convolution and residual concatenation was introduced light UNet to reduce the computational cost. Further, a robust segmentation loss was applied to improve the performance. In the inference phase, only a light UNet is used, which required low time cost and less GPU memory utilization. The average DSC of this method in the validation set is 0.8718. The code is available in https://github.com/laihaoran/Semi-SupervisednnUNet.
Semi-Supervised Hybrid Spine Network for Segmentation of Spine MR Images
Mar 23, 2022



Abstract:Automatic segmentation of vertebral bodies (VBs) and intervertebral discs (IVDs) in 3D magnetic resonance (MR) images is vital in diagnosing and treating spinal diseases. However, segmenting the VBs and IVDs simultaneously is not trivial. Moreover, problems exist, including blurry segmentation caused by anisotropy resolution, high computational cost, inter-class similarity and intra-class variability, and data imbalances. We proposed a two-stage algorithm, named semi-supervised hybrid spine network (SSHSNet), to address these problems by achieving accurate simultaneous VB and IVD segmentation. In the first stage, we constructed a 2D semi-supervised DeepLabv3+ by using cross pseudo supervision to obtain intra-slice features and coarse segmentation. In the second stage, a 3D full-resolution patch-based DeepLabv3+ was built. This model can be used to extract inter-slice information and combine the coarse segmentation and intra-slice features provided from the first stage. Moreover, a cross tri-attention module was applied to compensate for the loss of inter-slice and intra-slice information separately generated from 2D and 3D networks, thereby improving feature representation ability and achieving satisfactory segmentation results. The proposed SSHSNet was validated on a publicly available spine MR image dataset, and remarkable segmentation performance was achieved. Moreover, results show that the proposed method has great potential in dealing with the data imbalance problem. Based on previous reports, few studies have incorporated a semi-supervised learning strategy with a cross attention mechanism for spine segmentation. Therefore, the proposed method may provide a useful tool for spine segmentation and aid clinically in spinal disease diagnoses and treatments. Codes are publicly available at: https://github.com/Meiyan88/SSHSNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge