Oisin Mac Aodha
MotionPhysics: Learnable Motion Distillation for Text-Guided Simulation
Jan 01, 2026Abstract:Accurately simulating existing 3D objects and a wide variety of materials often demands expert knowledge and time-consuming physical parameter tuning to achieve the desired dynamic behavior. We introduce MotionPhysics, an end-to-end differentiable framework that infers plausible physical parameters from a user-provided natural language prompt for a chosen 3D scene of interest, removing the need for guidance from ground-truth trajectories or annotated videos. Our approach first utilizes a multimodal large language model to estimate material parameter values, which are constrained to lie within plausible ranges. We further propose a learnable motion distillation loss that extracts robust motion priors from pretrained video diffusion models while minimizing appearance and geometry inductive biases to guide the simulation. We evaluate MotionPhysics across more than thirty scenarios, including real-world, human-designed, and AI-generated 3D objects, spanning a wide range of materials such as elastic solids, metals, foams, sand, and both Newtonian and non-Newtonian fluids. We demonstrate that MotionPhysics produces visually realistic dynamic simulations guided by natural language, surpassing the state of the art while automatically determining physically plausible parameters. The code and project page are available at: https://wangmiaowei.github.io/MotionPhysics.github.io/.
INQUIRE-Search: A Framework for Interactive Discovery in Large-Scale Biodiversity Databases
Nov 19, 2025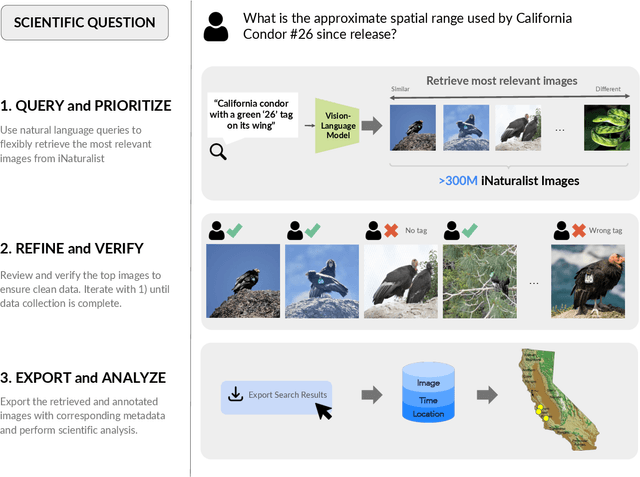
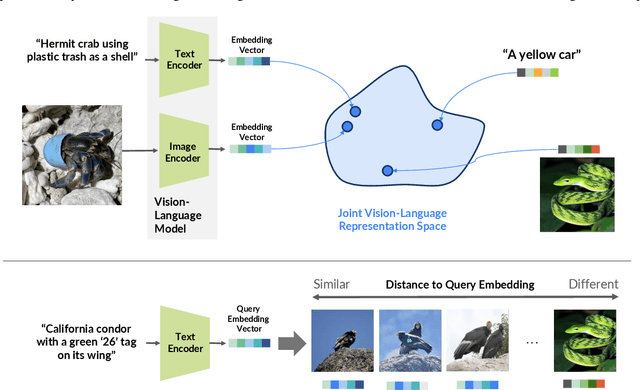

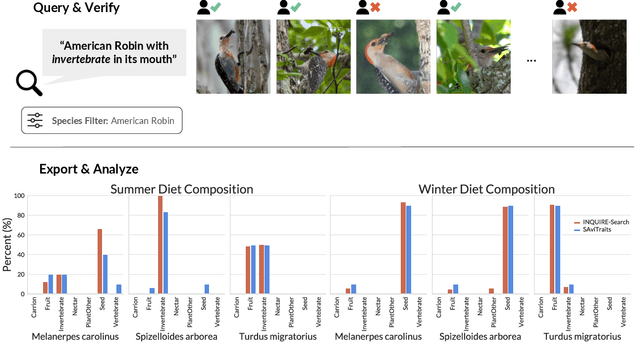
Abstract:Large community science platforms such as iNaturalist contain hundreds of millions of biodiversity images that often capture ecological context on behaviors, interactions, phenology, and habitat. Yet most ecological workflows rely on metadata filtering or manual inspection, leaving this secondary information inaccessible at scale. We introduce INQUIRE-Search, an open-source system that enables scientists to rapidly and interactively search within an ecological image database for specific concepts using natural language, verify and export relevant observations, and utilize this discovered data for novel scientific analysis. Compared to traditional methods, INQUIRE-Search takes a fraction of the time, opening up new possibilities for scientific questions that can be explored. Through five case studies, we show the diversity of scientific applications that a tool like INQUIRE-Search can support, from seasonal variation in behavior across species to forest regrowth after wildfires. These examples demonstrate a new paradigm for interactive, efficient, and scalable scientific discovery that can begin to unlock previously inaccessible scientific value in large-scale biodiversity datasets. Finally, we emphasize using such AI-enabled discovery tools for science call for experts to reframe the priorities of the scientific process and develop novel methods for experiment design, data collection, survey effort, and uncertainty analysis.
Attentive Feature Aggregation or: How Policies Learn to Stop Worrying about Robustness and Attend to Task-Relevant Visual Cues
Nov 13, 2025Abstract:The adoption of pre-trained visual representations (PVRs), leveraging features from large-scale vision models, has become a popular paradigm for training visuomotor policies. However, these powerful representations can encode a broad range of task-irrelevant scene information, making the resulting trained policies vulnerable to out-of-domain visual changes and distractors. In this work we address visuomotor policy feature pooling as a solution to the observed lack of robustness in perturbed scenes. We achieve this via Attentive Feature Aggregation (AFA), a lightweight, trainable pooling mechanism that learns to naturally attend to task-relevant visual cues, ignoring even semantically rich scene distractors. Through extensive experiments in both simulation and the real world, we demonstrate that policies trained with AFA significantly outperform standard pooling approaches in the presence of visual perturbations, without requiring expensive dataset augmentation or fine-tuning of the PVR. Our findings show that ignoring extraneous visual information is a crucial step towards deploying robust and generalisable visuomotor policies. Project Page: tsagkas.github.io/afa
CleverBirds: A Multiple-Choice Benchmark for Fine-grained Human Knowledge Tracing
Nov 11, 2025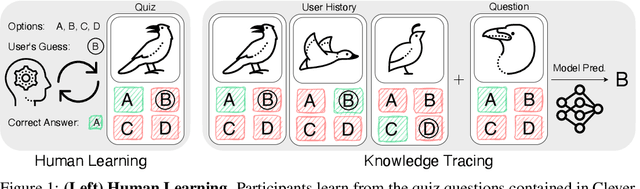

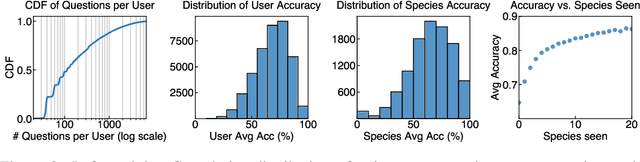
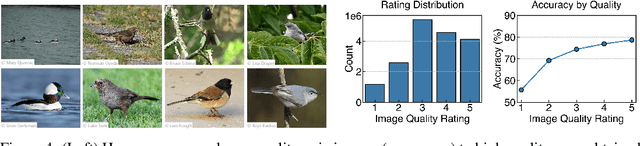
Abstract:Mastering fine-grained visual recognition, essential in many expert domains, can require that specialists undergo years of dedicated training. Modeling the progression of such expertize in humans remains challenging, and accurately inferring a human learner's knowledge state is a key step toward understanding visual learning. We introduce CleverBirds, a large-scale knowledge tracing benchmark for fine-grained bird species recognition. Collected by the citizen-science platform eBird, it offers insight into how individuals acquire expertize in complex fine-grained classification. More than 40,000 participants have engaged in the quiz, answering over 17 million multiple-choice questions spanning over 10,000 bird species, with long-range learning patterns across an average of 400 questions per participant. We release this dataset to support the development and evaluation of new methods for visual knowledge tracing. We show that tracking learners' knowledge is challenging, especially across participant subgroups and question types, with different forms of contextual information offering varying degrees of predictive benefit. CleverBirds is among the largest benchmark of its kind, offering a substantially higher number of learnable concepts. With it, we hope to enable new avenues for studying the development of visual expertize over time and across individuals.
Enhancing Tactile-based Reinforcement Learning for Robotic Control
Oct 24, 2025Abstract:Achieving safe, reliable real-world robotic manipulation requires agents to evolve beyond vision and incorporate tactile sensing to overcome sensory deficits and reliance on idealised state information. Despite its potential, the efficacy of tactile sensing in reinforcement learning (RL) remains inconsistent. We address this by developing self-supervised learning (SSL) methodologies to more effectively harness tactile observations, focusing on a scalable setup of proprioception and sparse binary contacts. We empirically demonstrate that sparse binary tactile signals are critical for dexterity, particularly for interactions that proprioceptive control errors do not register, such as decoupled robot-object motions. Our agents achieve superhuman dexterity in complex contact tasks (ball bouncing and Baoding ball rotation). Furthermore, we find that decoupling the SSL memory from the on-policy memory can improve performance. We release the Robot Tactile Olympiad (RoTO) benchmark to standardise and promote future research in tactile-based manipulation. Project page: https://elle-miller.github.io/tactile_rl
Sample-efficient Integration of New Modalities into Large Language Models
Sep 04, 2025Abstract:Multimodal foundation models can process several modalities. However, since the space of possible modalities is large and evolving over time, training a model from scratch to encompass all modalities is unfeasible. Moreover, integrating a modality into a pre-existing foundation model currently requires a significant amount of paired data, which is often not available for low-resource modalities. In this paper, we introduce a method for sample-efficient modality integration (SEMI) into Large Language Models (LLMs). To this end, we devise a hypernetwork that can adapt a shared projector -- placed between modality-specific encoders and an LLM -- to any modality. The hypernetwork, trained on high-resource modalities (i.e., text, speech, audio, video), is conditioned on a few samples from any arbitrary modality at inference time to generate a suitable adapter. To increase the diversity of training modalities, we artificially multiply the number of encoders through isometric transformations. We find that SEMI achieves a significant boost in sample efficiency during few-shot integration of new modalities (i.e., satellite images, astronomical images, inertial measurements, and molecules) with encoders of arbitrary embedding dimensionality. For instance, to reach the same accuracy as 32-shot SEMI, training the projector from scratch needs 64$\times$ more data. As a result, SEMI holds promise to extend the modality coverage of foundation models.
Interpretable Text-Guided Image Clustering via Iterative Search
Jun 14, 2025



Abstract:Traditional clustering methods aim to group unlabeled data points based on their similarity to each other. However, clustering, in the absence of additional information, is an ill-posed problem as there may be many different, yet equally valid, ways to partition a dataset. Distinct users may want to use different criteria to form clusters in the same data, e.g. shape v.s. color. Recently introduced text-guided image clustering methods aim to address this ambiguity by allowing users to specify the criteria of interest using natural language instructions. This instruction provides the necessary context and control needed to obtain clusters that are more aligned with the users' intent. We propose a new text-guided clustering approach named ITGC that uses an iterative discovery process, guided by an unsupervised clustering objective, to generate interpretable visual concepts that better capture the criteria expressed in a user's instructions. We report superior performance compared to existing methods across a wide variety of image clustering and fine-grained classification benchmarks.
Jamais Vu: Exposing the Generalization Gap in Supervised Semantic Correspondence
Jun 09, 2025Abstract:Semantic correspondence (SC) aims to establish semantically meaningful matches across different instances of an object category. We illustrate how recent supervised SC methods remain limited in their ability to generalize beyond sparsely annotated training keypoints, effectively acting as keypoint detectors. To address this, we propose a novel approach for learning dense correspondences by lifting 2D keypoints into a canonical 3D space using monocular depth estimation. Our method constructs a continuous canonical manifold that captures object geometry without requiring explicit 3D supervision or camera annotations. Additionally, we introduce SPair-U, an extension of SPair-71k with novel keypoint annotations, to better assess generalization. Experiments not only demonstrate that our model significantly outperforms supervised baselines on unseen keypoints, highlighting its effectiveness in learning robust correspondences, but that unsupervised baselines outperform supervised counterparts when generalized across different datasets.
Representational Difference Explanations
May 29, 2025Abstract:We propose a method for discovering and visualizing the differences between two learned representations, enabling more direct and interpretable model comparisons. We validate our method, which we call Representational Differences Explanations (RDX), by using it to compare models with known conceptual differences and demonstrate that it recovers meaningful distinctions where existing explainable AI (XAI) techniques fail. Applied to state-of-the-art models on challenging subsets of the ImageNet and iNaturalist datasets, RDX reveals both insightful representational differences and subtle patterns in the data. Although comparison is a cornerstone of scientific analysis, current tools in machine learning, namely post hoc XAI methods, struggle to support model comparison effectively. Our work addresses this gap by introducing an effective and explainable tool for contrasting model representations.
MVSAnywhere: Zero-Shot Multi-View Stereo
Mar 28, 2025Abstract:Computing accurate depth from multiple views is a fundamental and longstanding challenge in computer vision. However, most existing approaches do not generalize well across different domains and scene types (e.g. indoor vs. outdoor). Training a general-purpose multi-view stereo model is challenging and raises several questions, e.g. how to best make use of transformer-based architectures, how to incorporate additional metadata when there is a variable number of input views, and how to estimate the range of valid depths which can vary considerably across different scenes and is typically not known a priori? To address these issues, we introduce MVSA, a novel and versatile Multi-View Stereo architecture that aims to work Anywhere by generalizing across diverse domains and depth ranges. MVSA combines monocular and multi-view cues with an adaptive cost volume to deal with scale-related issues. We demonstrate state-of-the-art zero-shot depth estimation on the Robust Multi-View Depth Benchmark, surpassing existing multi-view stereo and monocular baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge