Elijah Cole
Few-shot Species Range Estimation
Feb 20, 2025Abstract:Knowing where a particular species can or cannot be found on Earth is crucial for ecological research and conservation efforts. By mapping the spatial ranges of all species, we would obtain deeper insights into how global biodiversity is affected by climate change and habitat loss. However, accurate range estimates are only available for a relatively small proportion of all known species. For the majority of the remaining species, we often only have a small number of records denoting the spatial locations where they have previously been observed. We outline a new approach for few-shot species range estimation to address the challenge of accurately estimating the range of a species from limited data. During inference, our model takes a set of spatial locations as input, along with optional metadata such as text or an image, and outputs a species encoding that can be used to predict the range of a previously unseen species in feed-forward manner. We validate our method on two challenging benchmarks, where we obtain state-of-the-art range estimation performance, in a fraction of the compute time, compared to recent alternative approaches.
WildSAT: Learning Satellite Image Representations from Wildlife Observations
Dec 19, 2024Abstract:What does the presence of a species reveal about a geographic location? We posit that habitat, climate, and environmental preferences reflected in species distributions provide a rich source of supervision for learning satellite image representations. We introduce WildSAT, which pairs satellite images with millions of geo-tagged wildlife observations readily-available on citizen science platforms. WildSAT uses a contrastive learning framework to combine information from species distribution maps with text descriptions that capture habitat and range details, alongside satellite images, to train or fine-tune models. On a range of downstream satellite image recognition tasks, this significantly improves the performance of both randomly initialized models and pre-trained models from sources like ImageNet or specialized satellite image datasets. Additionally, the alignment with text enables zero-shot retrieval, allowing for search based on general descriptions of locations. We demonstrate that WildSAT achieves better representations than recent methods that utilize other forms of cross-modal supervision, such as aligning satellite images with ground images or wildlife photos. Finally, we analyze the impact of various design choices on downstream performance, highlighting the general applicability of our approach.
Combining Observational Data and Language for Species Range Estimation
Oct 14, 2024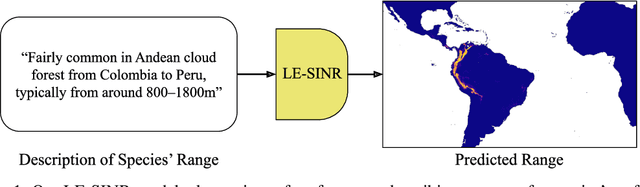
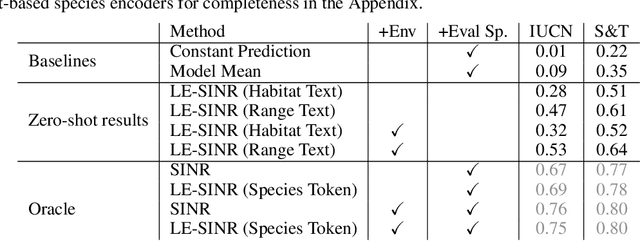
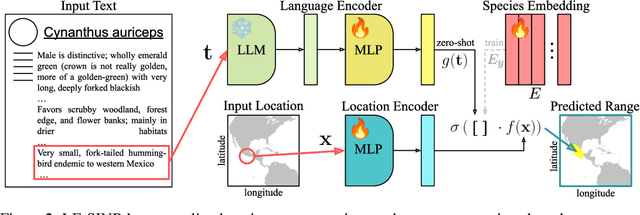
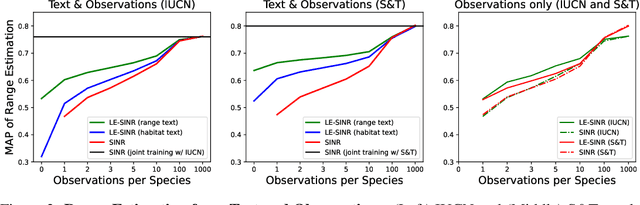
Abstract:Species range maps (SRMs) are essential tools for research and policy-making in ecology, conservation, and environmental management. However, traditional SRMs rely on the availability of environmental covariates and high-quality species location observation data, both of which can be challenging to obtain due to geographic inaccessibility and resource constraints. We propose a novel approach combining millions of citizen science species observations with textual descriptions from Wikipedia, covering habitat preferences and range descriptions for tens of thousands of species. Our framework maps locations, species, and text descriptions into a common space, facilitating the learning of rich spatial covariates at a global scale and enabling zero-shot range estimation from textual descriptions. Evaluated on held-out species, our zero-shot SRMs significantly outperform baselines and match the performance of SRMs obtained using tens of observations. Our approach also acts as a strong prior when combined with observational data, resulting in more accurate range estimation with less data. We present extensive quantitative and qualitative analyses of the learned representations in the context of range estimation and other spatial tasks, demonstrating the effectiveness of our approach.
A Closer Look at Benchmarking Self-Supervised Pre-training with Image Classification
Jul 18, 2024



Abstract:Self-supervised learning (SSL) is a machine learning approach where the data itself provides supervision, eliminating the need for external labels. The model is forced to learn about the data structure or context by solving a pretext task. With SSL, models can learn from abundant and cheap unlabeled data, significantly reducing the cost of training models where labels are expensive or inaccessible. In Computer Vision, SSL is widely used as pre-training followed by a downstream task, such as supervised transfer, few-shot learning on smaller labeled data sets, and/or unsupervised clustering. Unfortunately, it is infeasible to evaluate SSL methods on all possible downstream tasks and objectively measure the quality of the learned representation. Instead, SSL methods are evaluated using in-domain evaluation protocols, such as fine-tuning, linear probing, and k-nearest neighbors (kNN). However, it is not well understood how well these evaluation protocols estimate the representation quality of a pre-trained model for different downstream tasks under different conditions, such as dataset, metric, and model architecture. We study how classification-based evaluation protocols for SSL correlate and how well they predict downstream performance on different dataset types. Our study includes eleven common image datasets and 26 models that were pre-trained with different SSL methods or have different model backbones. We find that in-domain linear/kNN probing protocols are, on average, the best general predictors for out-of-domain performance. We further investigate the importance of batch normalization and evaluate how robust correlations are for different kinds of dataset domain shifts. We challenge assumptions about the relationship between discriminative and generative self-supervised methods, finding that most of their performance differences can be explained by changes to model backbones.
BenthIQ: a Transformer-Based Benthic Classification Model for Coral Restoration
Nov 22, 2023Abstract:Coral reefs are vital for marine biodiversity, coastal protection, and supporting human livelihoods globally. However, they are increasingly threatened by mass bleaching events, pollution, and unsustainable practices with the advent of climate change. Monitoring the health of these ecosystems is crucial for effective restoration and management. Current methods for creating benthic composition maps often compromise between spatial coverage and resolution. In this paper, we introduce BenthIQ, a multi-label semantic segmentation network designed for high-precision classification of underwater substrates, including live coral, algae, rock, and sand. Although commonly deployed CNNs are limited in learning long-range semantic information, transformer-based models have recently achieved state-of-the-art performance in vision tasks such as object detection and image classification. We integrate the hierarchical Swin Transformer as the backbone of a U-shaped encoder-decoder architecture for local-global semantic feature learning. Using a real-world case study in French Polynesia, we demonstrate that our approach outperforms traditional CNN and attention-based models on pixel-wise classification of shallow reef imagery.
Active Learning-Based Species Range Estimation
Nov 03, 2023Abstract:We propose a new active learning approach for efficiently estimating the geographic range of a species from a limited number of on the ground observations. We model the range of an unmapped species of interest as the weighted combination of estimated ranges obtained from a set of different species. We show that it is possible to generate this candidate set of ranges by using models that have been trained on large weakly supervised community collected observation data. From this, we develop a new active querying approach that sequentially selects geographic locations to visit that best reduce our uncertainty over an unmapped species' range. We conduct a detailed evaluation of our approach and compare it to existing active learning methods using an evaluation dataset containing expert-derived ranges for one thousand species. Our results demonstrate that our method outperforms alternative active learning methods and approaches the performance of end-to-end trained models, even when only using a fraction of the data. This highlights the utility of active learning via transfer learned spatial representations for species range estimation. It also emphasizes the value of leveraging emerging large-scale crowdsourced datasets, not only for modeling a species' range, but also for actively discovering them.
Spatial Implicit Neural Representations for Global-Scale Species Mapping
Jun 05, 2023Abstract:Estimating the geographical range of a species from sparse observations is a challenging and important geospatial prediction problem. Given a set of locations where a species has been observed, the goal is to build a model to predict whether the species is present or absent at any location. This problem has a long history in ecology, but traditional methods struggle to take advantage of emerging large-scale crowdsourced datasets which can include tens of millions of records for hundreds of thousands of species. In this work, we use Spatial Implicit Neural Representations (SINRs) to jointly estimate the geographical range of 47k species simultaneously. We find that our approach scales gracefully, making increasingly better predictions as we increase the number of species and the amount of data per species when training. To make this problem accessible to machine learning researchers, we provide four new benchmarks that measure different aspects of species range estimation and spatial representation learning. Using these benchmarks, we demonstrate that noisy and biased crowdsourced data can be combined with implicit neural representations to approximate expert-developed range maps for many species.
Understanding Label Bias in Single Positive Multi-Label Learning
May 24, 2023Abstract:Annotating data for multi-label classification is prohibitively expensive because every category of interest must be confirmed to be present or absent. Recent work on single positive multi-label (SPML) learning shows that it is possible to train effective multi-label classifiers using only one positive label per image. However, the standard benchmarks for SPML are derived from traditional multi-label classification datasets by retaining one positive label for each training example (chosen uniformly at random) and discarding all other labels. In realistic settings it is not likely that positive labels are chosen uniformly at random. This work introduces protocols for studying label bias in SPML and provides new empirical results.
Teaching Computer Vision for Ecology
Jan 05, 2023


Abstract:Computer vision can accelerate ecology research by automating the analysis of raw imagery from sensors like camera traps, drones, and satellites. However, computer vision is an emerging discipline that is rarely taught to ecologists. This work discusses our experience teaching a diverse group of ecologists to prototype and evaluate computer vision systems in the context of an intensive hands-on summer workshop. We explain the workshop structure, discuss common challenges, and propose best practices. This document is intended for computer scientists who teach computer vision across disciplines, but it may also be useful to ecologists or other domain experts who are learning to use computer vision themselves.
On Label Granularity and Object Localization
Jul 20, 2022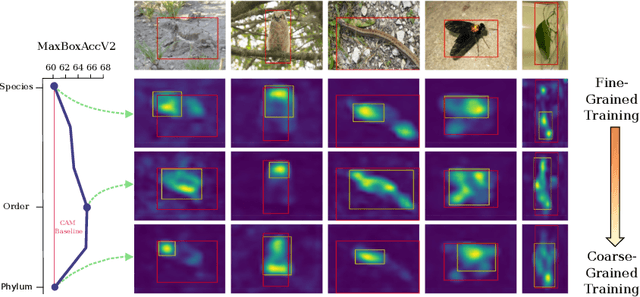
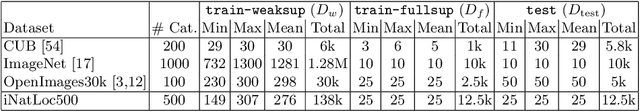
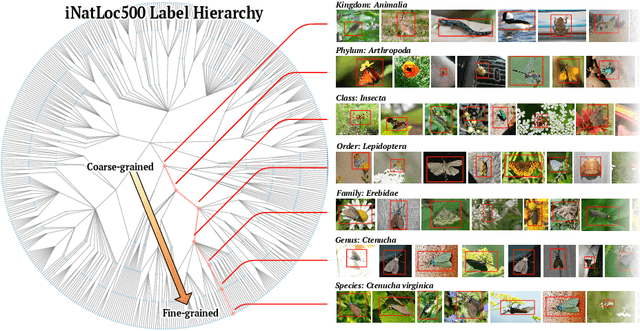
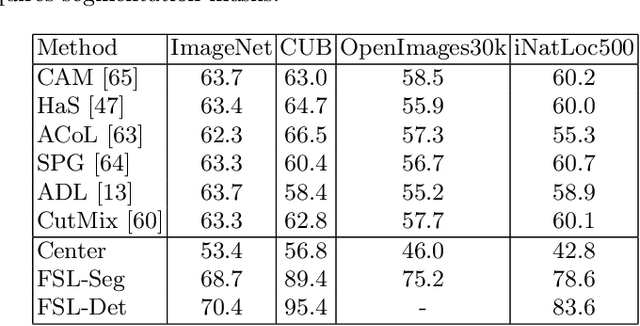
Abstract:Weakly supervised object localization (WSOL) aims to learn representations that encode object location using only image-level category labels. However, many objects can be labeled at different levels of granularity. Is it an animal, a bird, or a great horned owl? Which image-level labels should we use? In this paper we study the role of label granularity in WSOL. To facilitate this investigation we introduce iNatLoc500, a new large-scale fine-grained benchmark dataset for WSOL. Surprisingly, we find that choosing the right training label granularity provides a much larger performance boost than choosing the best WSOL algorithm. We also show that changing the label granularity can significantly improve data efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge