Mariano Cabezas
Enhancing Angular Resolution via Directionality Encoding and Geometric Constraints in Brain Diffusion Tensor Imaging
Sep 11, 2024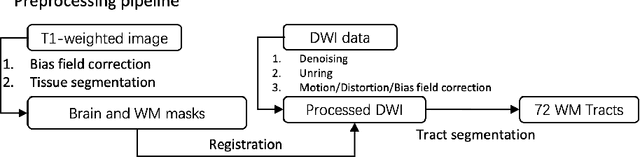

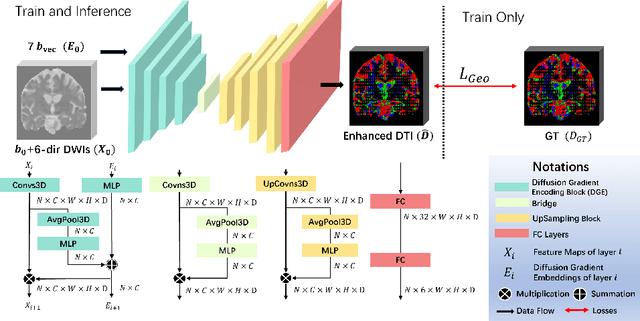

Abstract:Diffusion-weighted imaging (DWI) is a type of Magnetic Resonance Imaging (MRI) technique sensitised to the diffusivity of water molecules, offering the capability to inspect tissue microstructures and is the only in-vivo method to reconstruct white matter fiber tracts non-invasively. The DWI signal can be analysed with the diffusion tensor imaging (DTI) model to estimate the directionality of water diffusion within voxels. Several scalar metrics, including axial diffusivity (AD), mean diffusivity (MD), radial diffusivity (RD), and fractional anisotropy (FA), can be further derived from DTI to quantitatively summarise the microstructural integrity of brain tissue. These scalar metrics have played an important role in understanding the organisation and health of brain tissue at a microscopic level in clinical studies. However, reliable DTI metrics rely on DWI acquisitions with high gradient directions, which often go beyond the commonly used clinical protocols. To enhance the utility of clinically acquired DWI and save scanning time for robust DTI analysis, this work proposes DirGeo-DTI, a deep learning-based method to estimate reliable DTI metrics even from a set of DWIs acquired with the minimum theoretical number (6) of gradient directions. DirGeo-DTI leverages directional encoding and geometric constraints to facilitate the training process. Two public DWI datasets were used for evaluation, demonstrating the effectiveness of the proposed method. Extensive experimental results show that the proposed method achieves the best performance compared to existing DTI enhancement methods and potentially reveals further clinical insights with routine clinical DWI scans.
A benchmark for 2D foetal brain ultrasound analysis
Jun 25, 2024Abstract:Brain development involves a sequence of structural changes from early stages of the embryo until several months after birth. Currently, ultrasound is the established technique for screening due to its ability to acquire dynamic images in real-time without radiation and to its cost-efficiency. However, identifying abnormalities remains challenging due to the difficulty in interpreting foetal brain images. In this work we present a set of 104 2D foetal brain ultrasound images acquired during the 20th week of gestation that have been co-registered to a common space from a rough skull segmentation. The images are provided both on the original space and template space centred on the ellipses of all the subjects. Furthermore, the images have been annotated to highlight landmark points from structures of interest to analyse brain development. Both the final atlas template with probabilistic maps and the original images can be used to develop new segmentation techniques, test registration approaches for foetal brain ultrasound, extend our work to longitudinal datasets and to detect anomalies in new images.
How Much Data are Enough? Investigating Dataset Requirements for Patch-Based Brain MRI Segmentation Tasks
Apr 04, 2024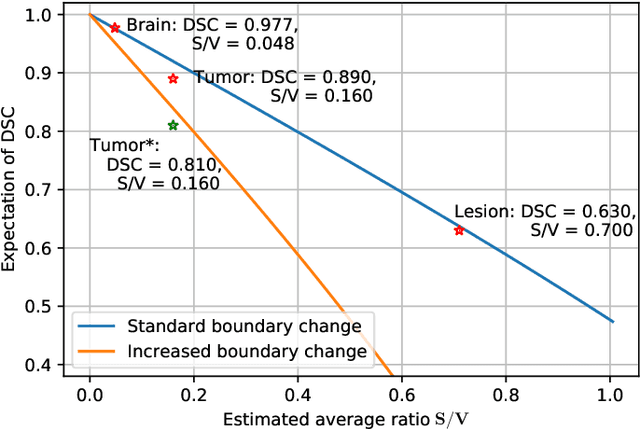
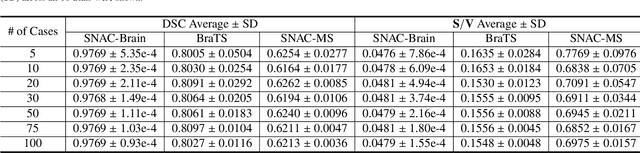

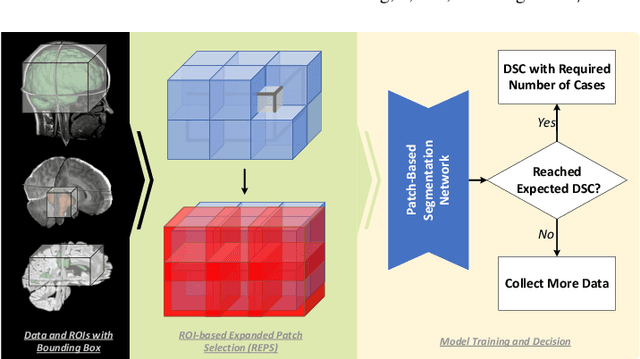
Abstract:Training deep neural networks reliably requires access to large-scale datasets. However, obtaining such datasets can be challenging, especially in the context of neuroimaging analysis tasks, where the cost associated with image acquisition and annotation can be prohibitive. To mitigate both the time and financial costs associated with model development, a clear understanding of the amount of data required to train a satisfactory model is crucial. This paper focuses on an early stage phase of deep learning research, prior to model development, and proposes a strategic framework for estimating the amount of annotated data required to train patch-based segmentation networks. This framework includes the establishment of performance expectations using a novel Minor Boundary Adjustment for Threshold (MinBAT) method, and standardizing patch selection through the ROI-based Expanded Patch Selection (REPS) method. Our experiments demonstrate that tasks involving regions of interest (ROIs) with different sizes or shapes may yield variably acceptable Dice Similarity Coefficient (DSC) scores. By setting an acceptable DSC as the target, the required amount of training data can be estimated and even predicted as data accumulates. This approach could assist researchers and engineers in estimating the cost associated with data collection and annotation when defining a new segmentation task based on deep neural networks, ultimately contributing to their efficient translation to real-world applications.
Improving Multiple Sclerosis Lesion Segmentation Across Clinical Sites: A Federated Learning Approach with Noise-Resilient Training
Aug 31, 2023



Abstract:Accurately measuring the evolution of Multiple Sclerosis (MS) with magnetic resonance imaging (MRI) critically informs understanding of disease progression and helps to direct therapeutic strategy. Deep learning models have shown promise for automatically segmenting MS lesions, but the scarcity of accurately annotated data hinders progress in this area. Obtaining sufficient data from a single clinical site is challenging and does not address the heterogeneous need for model robustness. Conversely, the collection of data from multiple sites introduces data privacy concerns and potential label noise due to varying annotation standards. To address this dilemma, we explore the use of the federated learning framework while considering label noise. Our approach enables collaboration among multiple clinical sites without compromising data privacy under a federated learning paradigm that incorporates a noise-robust training strategy based on label correction. Specifically, we introduce a Decoupled Hard Label Correction (DHLC) strategy that considers the imbalanced distribution and fuzzy boundaries of MS lesions, enabling the correction of false annotations based on prediction confidence. We also introduce a Centrally Enhanced Label Correction (CELC) strategy, which leverages the aggregated central model as a correction teacher for all sites, enhancing the reliability of the correction process. Extensive experiments conducted on two multi-site datasets demonstrate the effectiveness and robustness of our proposed methods, indicating their potential for clinical applications in multi-site collaborations.
Precise Few-shot Fat-free Thigh Muscle Segmentation in T1-weighted MRI
Apr 27, 2023Abstract:Precise thigh muscle volumes are crucial to monitor the motor functionality of patients with diseases that may result in various degrees of thigh muscle loss. T1-weighted MRI is the default surrogate to obtain thigh muscle masks due to its contrast between muscle and fat signals. Deep learning approaches have recently been widely used to obtain these masks through segmentation. However, due to the insufficient amount of precise annotations, thigh muscle masks generated by deep learning approaches tend to misclassify intra-muscular fat (IMF) as muscle impacting the analysis of muscle volumetrics. As IMF is infiltrated inside the muscle, human annotations require expertise and time. Thus, precise muscle masks where IMF is excluded are limited in practice. To alleviate this, we propose a few-shot segmentation framework to generate thigh muscle masks excluding IMF. In our framework, we design a novel pseudo-label correction and evaluation scheme, together with a new noise robust loss for exploiting high certainty areas. The proposed framework only takes $1\%$ of the fine-annotated training dataset, and achieves comparable performance with fully supervised methods according to the experimental results.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
TW-BAG: Tensor-wise Brain-aware Gate Network for Inpainting Disrupted Diffusion Tensor Imaging
Oct 31, 2022Abstract:Diffusion Weighted Imaging (DWI) is an advanced imaging technique commonly used in neuroscience and neurological clinical research through a Diffusion Tensor Imaging (DTI) model. Volumetric scalar metrics including fractional anisotropy, mean diffusivity, and axial diffusivity can be derived from the DTI model to summarise water diffusivity and other quantitative microstructural information for clinical studies. However, clinical practice constraints can lead to sub-optimal DWI acquisitions with missing slices (either due to a limited field of view or the acquisition of disrupted slices). To avoid discarding valuable subjects for group-wise studies, we propose a novel 3D Tensor-Wise Brain-Aware Gate network (TW-BAG) for inpainting disrupted DTIs. The proposed method is tailored to the problem with a dynamic gate mechanism and independent tensor-wise decoders. We evaluated the proposed method on the publicly available Human Connectome Project (HCP) dataset using common image similarity metrics derived from the predicted tensors and scalar DTI metrics. Our experimental results show that the proposed approach can reconstruct the original brain DTI volume and recover relevant clinical imaging information.
MS Lesion Segmentation: Revisiting Weighting Mechanisms for Federated Learning
May 03, 2022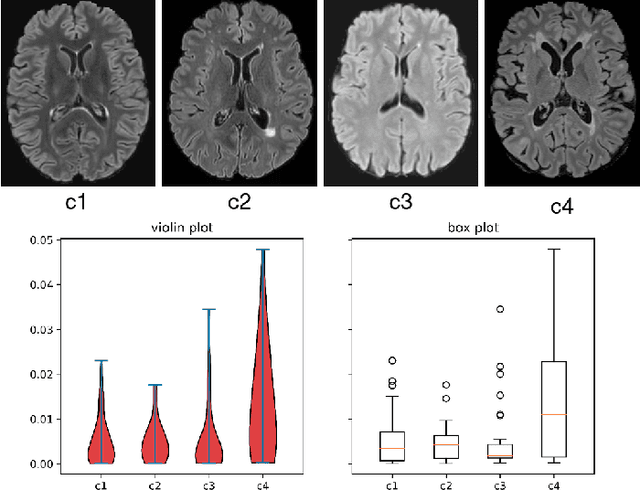
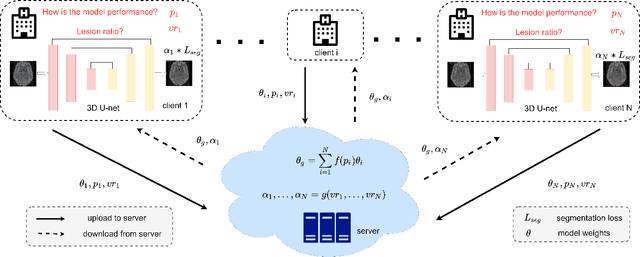
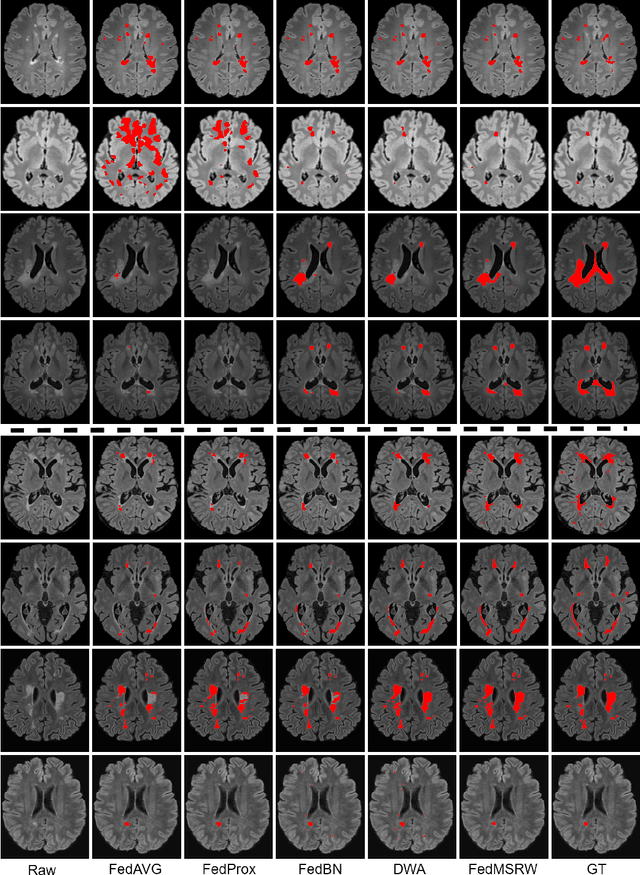
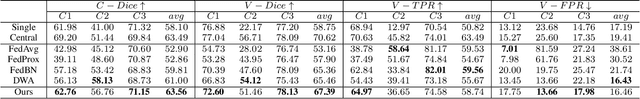
Abstract:Federated learning (FL) has been widely employed for medical image analysis to facilitate multi-client collaborative learning without sharing raw data. Despite great success, FL's performance is limited for multiple sclerosis (MS) lesion segmentation tasks, due to variance in lesion characteristics imparted by different scanners and acquisition parameters. In this work, we propose the first FL MS lesion segmentation framework via two effective re-weighting mechanisms. Specifically, a learnable weight is assigned to each local node during the aggregation process, based on its segmentation performance. In addition, the segmentation loss function in each client is also re-weighted according to the lesion volume for the data during training. Comparison experiments on two FL MS segmentation scenarios using public and clinical datasets have demonstrated the effectiveness of the proposed method by outperforming other FL methods significantly. Furthermore, the segmentation performance of FL incorporating our proposed aggregation mechanism can exceed centralised training with all the raw data. The extensive evaluation also indicated the superiority of our method when estimating brain volume differences estimation after lesion inpainting.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021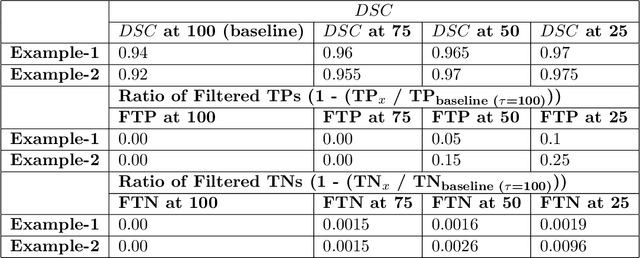
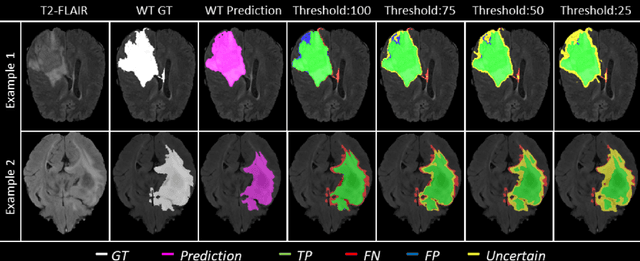
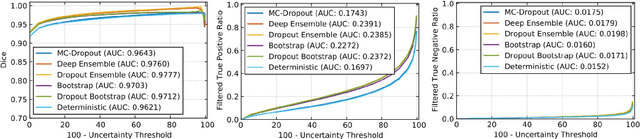
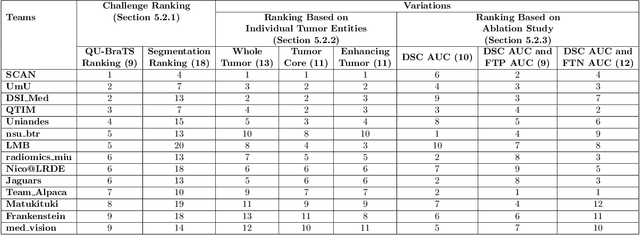
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Multiple Sclerosis Lesion Analysis in Brain Magnetic Resonance Images: Techniques and Clinical Applications
Apr 20, 2021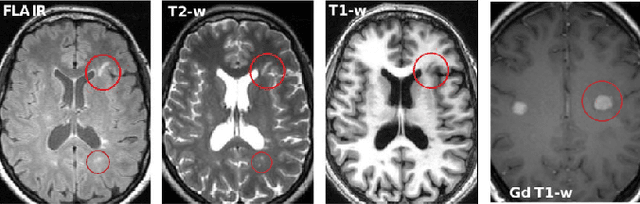
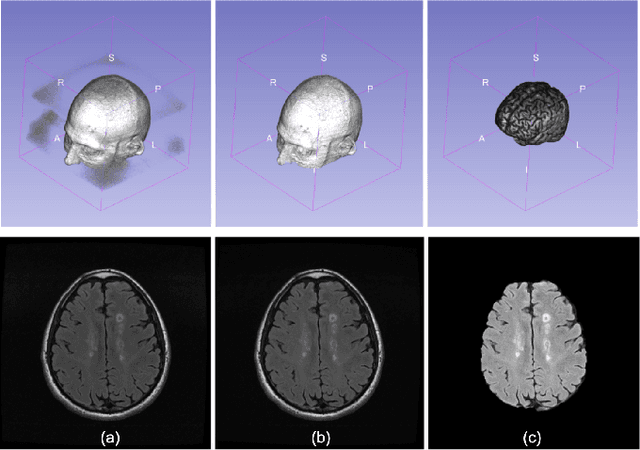
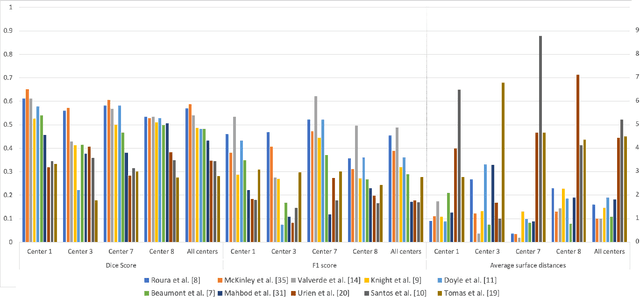
Abstract:Multiple sclerosis (MS) is a chronic inflammatory and degenerative disease of the central nervous system, characterized by the appearance of focal lesions in the white and gray matter that topographically correlate with an individual patient's neurological symptoms and signs. Magnetic resonance imaging (MRI) provides detailed in-vivo structural information, permitting the quantification and categorization of MS lesions that critically inform disease management. Traditionally, MS lesions have been manually annotated on 2D MRI slices, a process that is inefficient and prone to inter-/intra-observer errors. Recently, automated statistical imaging analysis techniques have been proposed to extract and segment MS lesions based on MRI voxel intensity. However, their effectiveness is limited by the heterogeneity of both MRI data acquisition techniques and the appearance of MS lesions. By learning complex lesion representations directly from images, deep learning techniques have achieved remarkable breakthroughs in the MS lesion segmentation task. Here, we provide a comprehensive review of state-of-the-art automatic statistical and deep-learning MS segmentation methods and discuss current and future clinical applications. Further, we review technical strategies, such as domain adaptation, to enhance MS lesion segmentation in real-world clinical settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge