Fernando Calamante
Age Sensitive Hippocampal Functional Connectivity: New Insights from 3D CNNs and Saliency Mapping
Jul 02, 2025Abstract:Grey matter loss in the hippocampus is a hallmark of neurobiological aging, yet understanding the corresponding changes in its functional connectivity remains limited. Seed-based functional connectivity (FC) analysis enables voxel-wise mapping of the hippocampus's synchronous activity with cortical regions, offering a window into functional reorganization during aging. In this study, we develop an interpretable deep learning framework to predict brain age from hippocampal FC using a three-dimensional convolutional neural network (3D CNN) combined with LayerCAM saliency mapping. This approach maps key hippocampal-cortical connections, particularly with the precuneus, cuneus, posterior cingulate cortex, parahippocampal cortex, left superior parietal lobule, and right superior temporal sulcus, that are highly sensitive to age. Critically, disaggregating anterior and posterior hippocampal FC reveals distinct mapping aligned with their known functional specializations. These findings provide new insights into the functional mechanisms of hippocampal aging and demonstrate the power of explainable deep learning to uncover biologically meaningful patterns in neuroimaging data.
Voxel-Level Brain States Prediction Using Swin Transformer
Jun 13, 2025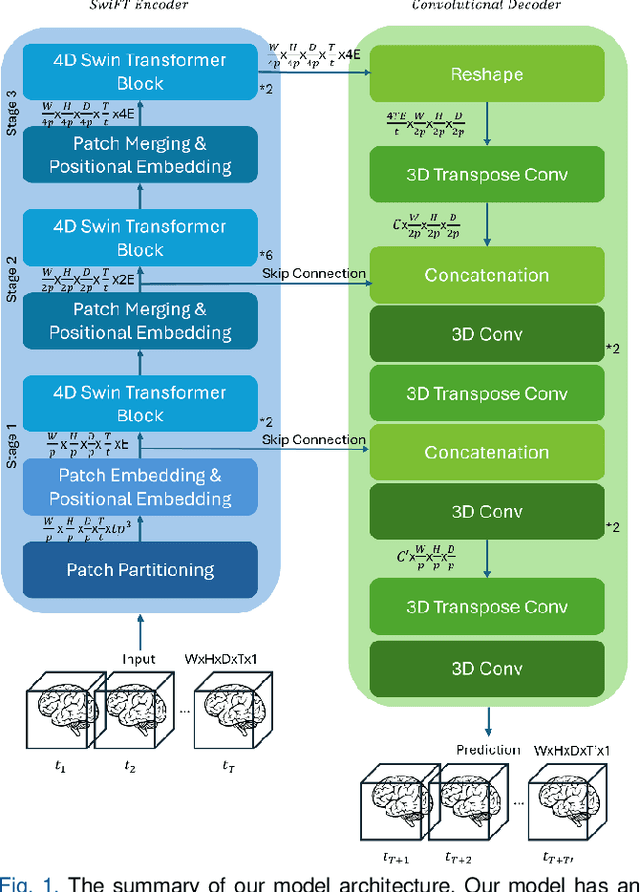
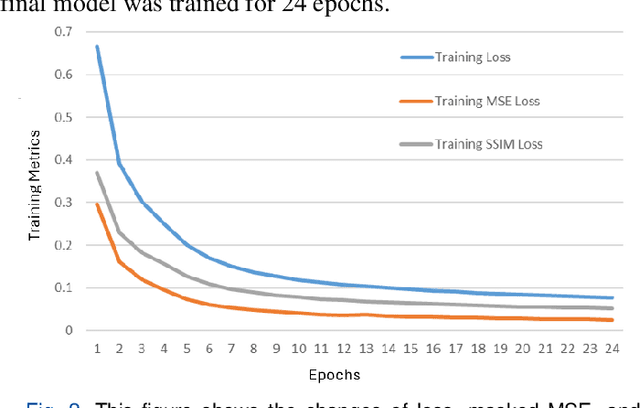
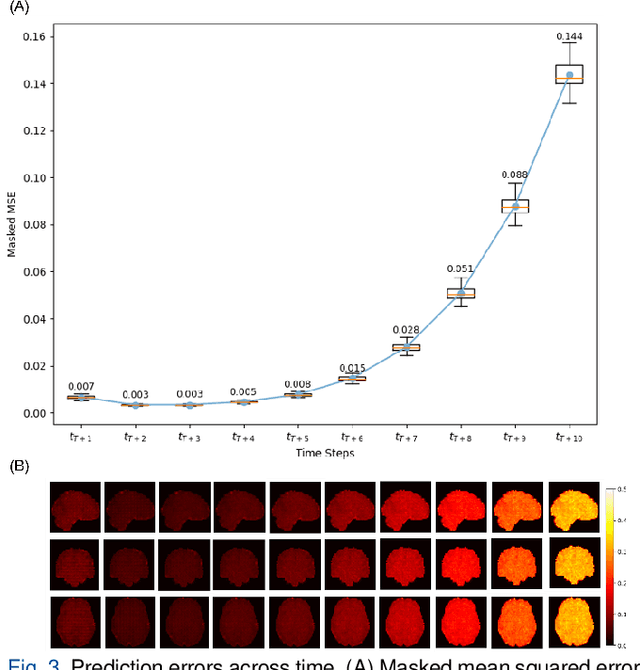
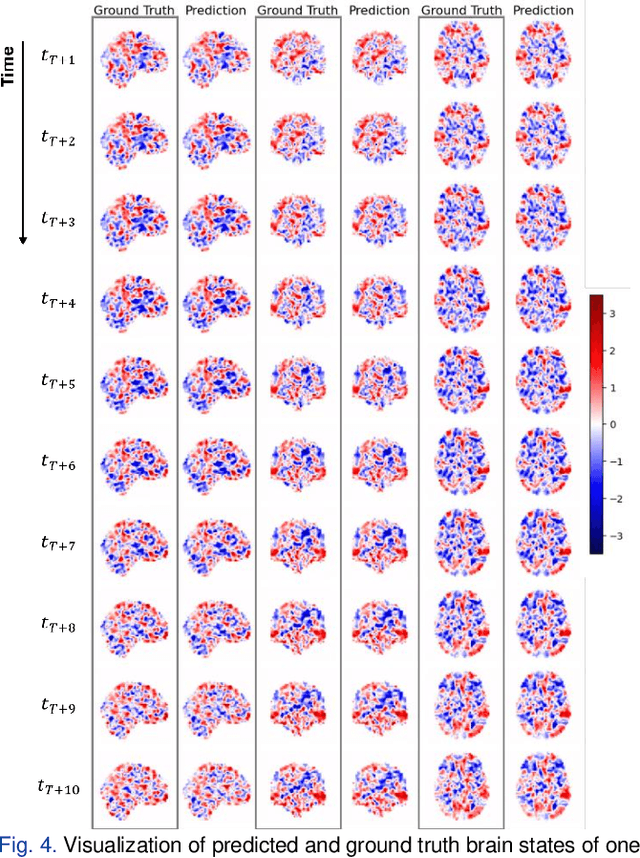
Abstract:Understanding brain dynamics is important for neuroscience and mental health. Functional magnetic resonance imaging (fMRI) enables the measurement of neural activities through blood-oxygen-level-dependent (BOLD) signals, which represent brain states. In this study, we aim to predict future human resting brain states with fMRI. Due to the 3D voxel-wise spatial organization and temporal dependencies of the fMRI data, we propose a novel architecture which employs a 4D Shifted Window (Swin) Transformer as encoder to efficiently learn spatio-temporal information and a convolutional decoder to enable brain state prediction at the same spatial and temporal resolution as the input fMRI data. We used 100 unrelated subjects from the Human Connectome Project (HCP) for model training and testing. Our novel model has shown high accuracy when predicting 7.2s resting-state brain activities based on the prior 23.04s fMRI time series. The predicted brain states highly resemble BOLD contrast and dynamics. This work shows promising evidence that the spatiotemporal organization of the human brain can be learned by a Swin Transformer model, at high resolution, which provides a potential for reducing the fMRI scan time and the development of brain-computer interfaces in the future.
Enhancing Angular Resolution via Directionality Encoding and Geometric Constraints in Brain Diffusion Tensor Imaging
Sep 11, 2024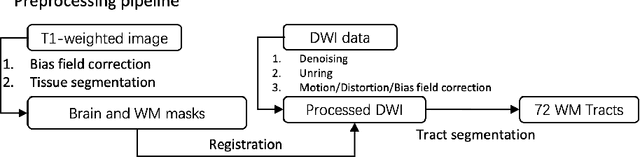

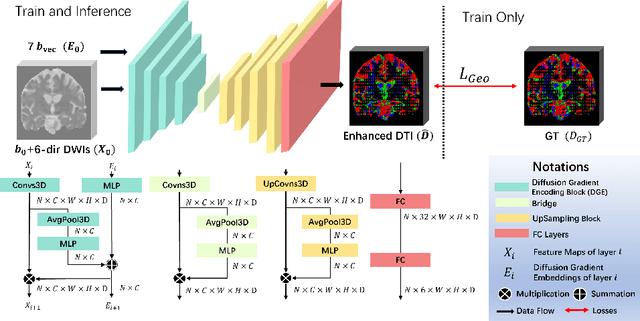

Abstract:Diffusion-weighted imaging (DWI) is a type of Magnetic Resonance Imaging (MRI) technique sensitised to the diffusivity of water molecules, offering the capability to inspect tissue microstructures and is the only in-vivo method to reconstruct white matter fiber tracts non-invasively. The DWI signal can be analysed with the diffusion tensor imaging (DTI) model to estimate the directionality of water diffusion within voxels. Several scalar metrics, including axial diffusivity (AD), mean diffusivity (MD), radial diffusivity (RD), and fractional anisotropy (FA), can be further derived from DTI to quantitatively summarise the microstructural integrity of brain tissue. These scalar metrics have played an important role in understanding the organisation and health of brain tissue at a microscopic level in clinical studies. However, reliable DTI metrics rely on DWI acquisitions with high gradient directions, which often go beyond the commonly used clinical protocols. To enhance the utility of clinically acquired DWI and save scanning time for robust DTI analysis, this work proposes DirGeo-DTI, a deep learning-based method to estimate reliable DTI metrics even from a set of DWIs acquired with the minimum theoretical number (6) of gradient directions. DirGeo-DTI leverages directional encoding and geometric constraints to facilitate the training process. Two public DWI datasets were used for evaluation, demonstrating the effectiveness of the proposed method. Extensive experimental results show that the proposed method achieves the best performance compared to existing DTI enhancement methods and potentially reveals further clinical insights with routine clinical DWI scans.
How Much Data are Enough? Investigating Dataset Requirements for Patch-Based Brain MRI Segmentation Tasks
Apr 04, 2024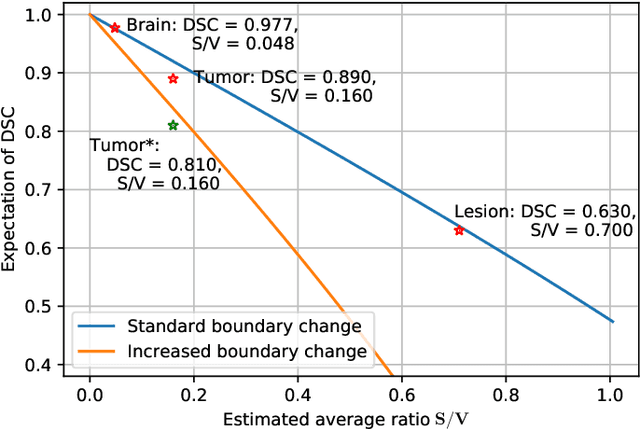
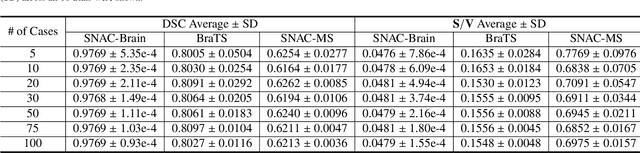

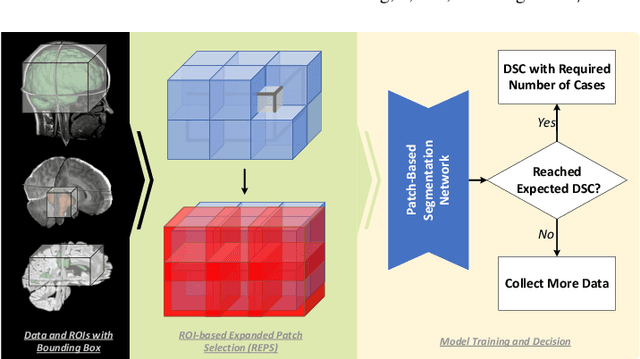
Abstract:Training deep neural networks reliably requires access to large-scale datasets. However, obtaining such datasets can be challenging, especially in the context of neuroimaging analysis tasks, where the cost associated with image acquisition and annotation can be prohibitive. To mitigate both the time and financial costs associated with model development, a clear understanding of the amount of data required to train a satisfactory model is crucial. This paper focuses on an early stage phase of deep learning research, prior to model development, and proposes a strategic framework for estimating the amount of annotated data required to train patch-based segmentation networks. This framework includes the establishment of performance expectations using a novel Minor Boundary Adjustment for Threshold (MinBAT) method, and standardizing patch selection through the ROI-based Expanded Patch Selection (REPS) method. Our experiments demonstrate that tasks involving regions of interest (ROIs) with different sizes or shapes may yield variably acceptable Dice Similarity Coefficient (DSC) scores. By setting an acceptable DSC as the target, the required amount of training data can be estimated and even predicted as data accumulates. This approach could assist researchers and engineers in estimating the cost associated with data collection and annotation when defining a new segmentation task based on deep neural networks, ultimately contributing to their efficient translation to real-world applications.
Improving Multiple Sclerosis Lesion Segmentation Across Clinical Sites: A Federated Learning Approach with Noise-Resilient Training
Aug 31, 2023



Abstract:Accurately measuring the evolution of Multiple Sclerosis (MS) with magnetic resonance imaging (MRI) critically informs understanding of disease progression and helps to direct therapeutic strategy. Deep learning models have shown promise for automatically segmenting MS lesions, but the scarcity of accurately annotated data hinders progress in this area. Obtaining sufficient data from a single clinical site is challenging and does not address the heterogeneous need for model robustness. Conversely, the collection of data from multiple sites introduces data privacy concerns and potential label noise due to varying annotation standards. To address this dilemma, we explore the use of the federated learning framework while considering label noise. Our approach enables collaboration among multiple clinical sites without compromising data privacy under a federated learning paradigm that incorporates a noise-robust training strategy based on label correction. Specifically, we introduce a Decoupled Hard Label Correction (DHLC) strategy that considers the imbalanced distribution and fuzzy boundaries of MS lesions, enabling the correction of false annotations based on prediction confidence. We also introduce a Centrally Enhanced Label Correction (CELC) strategy, which leverages the aggregated central model as a correction teacher for all sites, enhancing the reliability of the correction process. Extensive experiments conducted on two multi-site datasets demonstrate the effectiveness and robustness of our proposed methods, indicating their potential for clinical applications in multi-site collaborations.
MS Lesion Segmentation: Revisiting Weighting Mechanisms for Federated Learning
May 03, 2022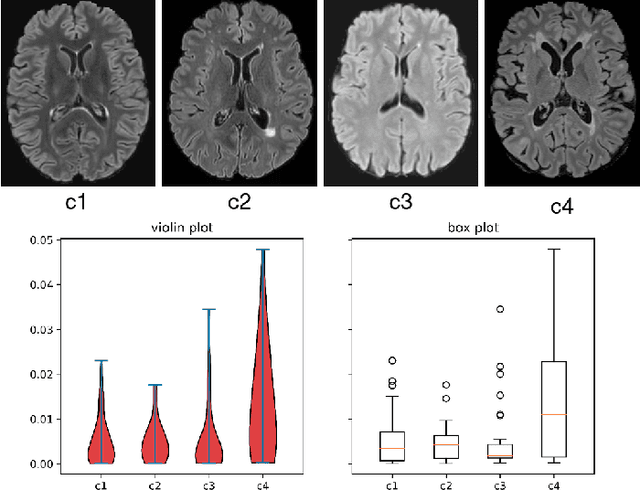
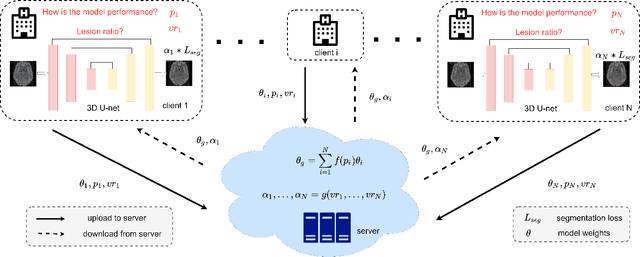
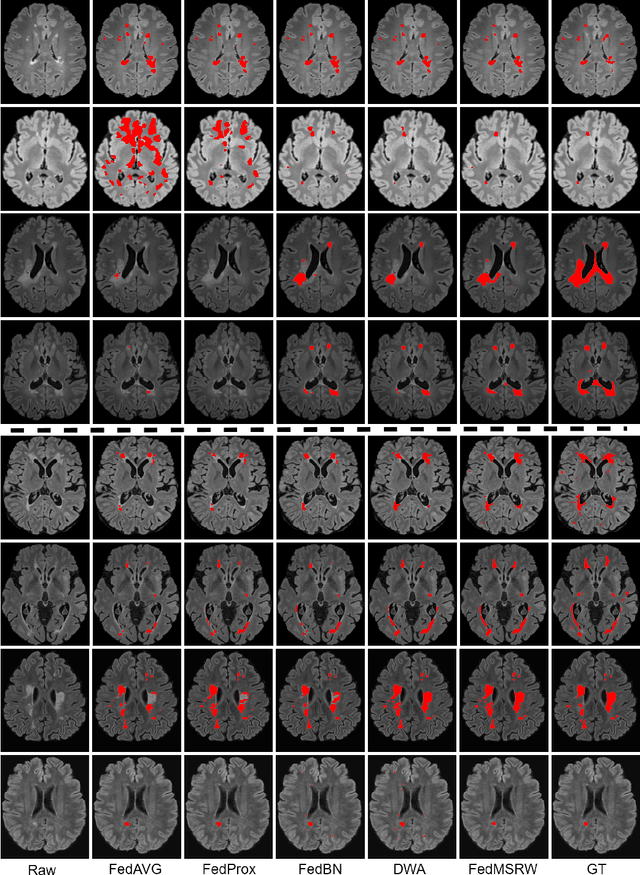
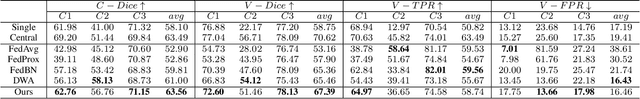
Abstract:Federated learning (FL) has been widely employed for medical image analysis to facilitate multi-client collaborative learning without sharing raw data. Despite great success, FL's performance is limited for multiple sclerosis (MS) lesion segmentation tasks, due to variance in lesion characteristics imparted by different scanners and acquisition parameters. In this work, we propose the first FL MS lesion segmentation framework via two effective re-weighting mechanisms. Specifically, a learnable weight is assigned to each local node during the aggregation process, based on its segmentation performance. In addition, the segmentation loss function in each client is also re-weighted according to the lesion volume for the data during training. Comparison experiments on two FL MS segmentation scenarios using public and clinical datasets have demonstrated the effectiveness of the proposed method by outperforming other FL methods significantly. Furthermore, the segmentation performance of FL incorporating our proposed aggregation mechanism can exceed centralised training with all the raw data. The extensive evaluation also indicated the superiority of our method when estimating brain volume differences estimation after lesion inpainting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge