Long-Kai Huang
Learning Where to Edit Vision Transformers
Nov 04, 2024



Abstract:Model editing aims to data-efficiently correct predictive errors of large pre-trained models while ensuring generalization to neighboring failures and locality to minimize unintended effects on unrelated examples. While significant progress has been made in editing Transformer-based large language models, effective strategies for editing vision Transformers (ViTs) in computer vision remain largely untapped. In this paper, we take initial steps towards correcting predictive errors of ViTs, particularly those arising from subpopulation shifts. Taking a locate-then-edit approach, we first address the where-to-edit challenge by meta-learning a hypernetwork on CutMix-augmented data generated for editing reliability. This trained hypernetwork produces generalizable binary masks that identify a sparse subset of structured model parameters, responsive to real-world failure samples. Afterward, we solve the how-to-edit problem by simply fine-tuning the identified parameters using a variant of gradient descent to achieve successful edits. To validate our method, we construct an editing benchmark that introduces subpopulation shifts towards natural underrepresented images and AI-generated images, thereby revealing the limitations of pre-trained ViTs for object recognition. Our approach not only achieves superior performance on the proposed benchmark but also allows for adjustable trade-offs between generalization and locality. Our code is available at https://github.com/hustyyq/Where-to-Edit.
Atomas: Hierarchical Alignment on Molecule-Text for Unified Molecule Understanding and Generation
Apr 23, 2024Abstract:Molecule-and-text cross-modal representation learning has emerged as a promising direction for enhancing the quality of molecular representation, thereby improving performance in various scientific fields, including drug discovery and materials science. Existing studies adopt a global alignment approach to learn the knowledge from different modalities. These global alignment approaches fail to capture fine-grained information, such as molecular fragments and their corresponding textual description, which is crucial for downstream tasks. Furthermore, it is incapable to model such information using a similar global alignment strategy due to data scarcity of paired local part annotated data from existing datasets. In this paper, we propose Atomas, a multi-modal molecular representation learning framework to jointly learn representations from SMILES string and text. We design a Hierarchical Adaptive Alignment model to concurrently learn the fine-grained fragment correspondence between two modalities and align these representations of fragments in three levels. Additionally, Atomas's end-to-end training framework incorporates the tasks of understanding and generating molecule, thereby supporting a wider range of downstream tasks. In the retrieval task, Atomas exhibits robust generalization ability and outperforms the baseline by 30.8% of recall@1 on average. In the generation task, Atomas achieves state-of-the-art results in both molecule captioning task and molecule generation task. Moreover, the visualization of the Hierarchical Adaptive Alignment model further confirms the chemical significance of our approach. Our codes can be found at https://anonymous.4open.science/r/Atomas-03C3.
Functional Protein Design with Local Domain Alignment
Apr 18, 2024



Abstract:The core challenge of de novo protein design lies in creating proteins with specific functions or properties, guided by certain conditions. Current models explore to generate protein using structural and evolutionary guidance, which only provide indirect conditions concerning functions and properties. However, textual annotations of proteins, especially the annotations for protein domains, which directly describe the protein's high-level functionalities, properties, and their correlation with target amino acid sequences, remain unexplored in the context of protein design tasks. In this paper, we propose Protein-Annotation Alignment Generation (PAAG), a multi-modality protein design framework that integrates the textual annotations extracted from protein database for controllable generation in sequence space. Specifically, within a multi-level alignment module, PAAG can explicitly generate proteins containing specific domains conditioned on the corresponding domain annotations, and can even design novel proteins with flexible combinations of different kinds of annotations. Our experimental results underscore the superiority of the aligned protein representations from PAAG over 7 prediction tasks. Furthermore, PAAG demonstrates a nearly sixfold increase in generation success rate (24.7% vs 4.7% in zinc finger, and 54.3% vs 8.7% in the immunoglobulin domain) in comparison to the existing model.
Invariant Test-Time Adaptation for Vision-Language Model Generalization
Mar 01, 2024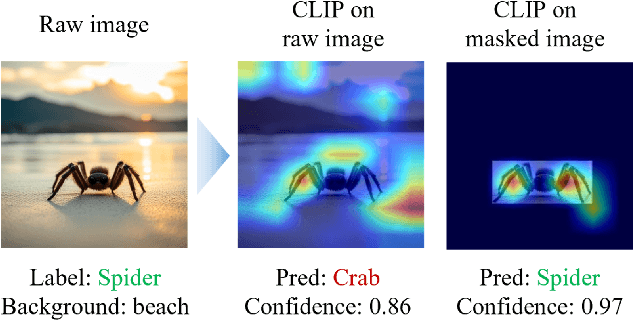

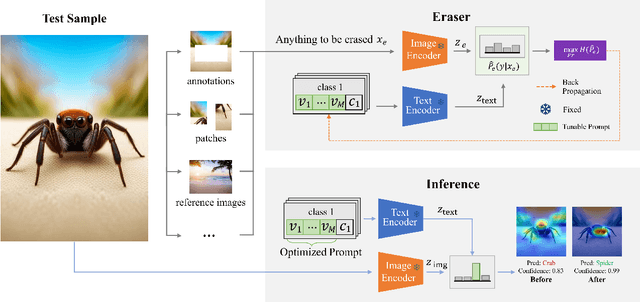

Abstract:Vision-language foundation models have exhibited remarkable success across a multitude of downstream tasks due to their scalability on extensive image-text paired datasets. However, these models display significant limitations when applied to long-tail tasks, such as fine-grained image classification, as a result of "decision shortcuts" that hinders their generalization capabilities. In this work, we find that the CLIP model possesses a rich set of features, encompassing both \textit{desired invariant causal features} and \textit{undesired decision shortcuts}. Moreover, the underperformance of CLIP on downstream tasks originates from its inability to effectively utilize pre-trained features in accordance with specific task requirements. To address this challenge, this paper introduces a test-time prompt tuning paradigm that optimizes a learnable prompt, thereby compelling the model to exploit genuine causal invariant features while disregarding decision shortcuts during the inference phase. The proposed method effectively alleviates excessive dependence on potentially misleading, task-irrelevant contextual information, while concurrently emphasizing critical, task-related visual cues. We conduct comparative analysis of the proposed method against various approaches which validates its effectiveness.
Concept-wise Fine-tuning Matters in Preventing Negative Transfer
Nov 12, 2023Abstract:A multitude of prevalent pre-trained models mark a major milestone in the development of artificial intelligence, while fine-tuning has been a common practice that enables pretrained models to figure prominently in a wide array of target datasets. Our empirical results reveal that off-the-shelf finetuning techniques are far from adequate to mitigate negative transfer caused by two types of underperforming features in a pre-trained model, including rare features and spuriously correlated features. Rooted in structural causal models of predictions after fine-tuning, we propose a Concept-wise fine-tuning (Concept-Tuning) approach which refines feature representations in the level of patches with each patch encoding a concept. Concept-Tuning minimizes the negative impacts of rare features and spuriously correlated features by (1) maximizing the mutual information between examples in the same category with regard to a slice of rare features (a patch) and (2) applying front-door adjustment via attention neural networks in channels and feature slices (patches). The proposed Concept-Tuning consistently and significantly (by up to 4.76%) improves prior state-of-the-art fine-tuning methods on eleven datasets, diverse pre-training strategies (supervised and self-supervised ones), various network architectures, and sample sizes in a target dataset.
Improving Generalizability of Graph Anomaly Detection Models via Data Augmentation
Jun 18, 2023Abstract:Graph anomaly detection (GAD) is a vital task since even a few anomalies can pose huge threats to benign users. Recent semi-supervised GAD methods, which can effectively leverage the available labels as prior knowledge, have achieved superior performances than unsupervised methods. In practice, people usually need to identify anomalies on new (sub)graphs to secure their business, but they may lack labels to train an effective detection model. One natural idea is to directly adopt a trained GAD model to the new (sub)graph for testing. However, we find that existing semi-supervised GAD methods suffer from poor generalization issue, i.e., well-trained models could not perform well on an unseen area (i.e., not accessible in training) of the same graph. It may cause great troubles. In this paper, we base on the phenomenon and propose a general and novel research problem of generalized graph anomaly detection that aims to effectively identify anomalies on both the training-domain graph and unseen testing graph to eliminate potential dangers. Nevertheless, it is a challenging task since only limited labels are available, and the normal background may differ between training and testing data. Accordingly, we propose a data augmentation method named \textit{AugAN} (\uline{Aug}mentation for \uline{A}nomaly and \uline{N}ormal distributions) to enrich training data and boost the generalizability of GAD models. Experiments verify the effectiveness of our method in improving model generalizability.
* Accepted to IEEE Transactions on Knowledge and Data Engineering (TKDE). arXiv admin note: substantial text overlap with arXiv:2209.10168
Learning to generate imaginary tasks for improving generalization in meta-learning
Jun 09, 2022



Abstract:The success of meta-learning on existing benchmarks is predicated on the assumption that the distribution of meta-training tasks covers meta-testing tasks. Frequent violation of the assumption in applications with either insufficient tasks or a very narrow meta-training task distribution leads to memorization or learner overfitting. Recent solutions have pursued augmentation of meta-training tasks, while it is still an open question to generate both correct and sufficiently imaginary tasks. In this paper, we seek an approach that up-samples meta-training tasks from the task representation via a task up-sampling network. Besides, the resulting approach named Adversarial Task Up-sampling (ATU) suffices to generate tasks that can maximally contribute to the latest meta-learner by maximizing an adversarial loss. On few-shot sine regression and image classification datasets, we empirically validate the marked improvement of ATU over state-of-the-art task augmentation strategies in the meta-testing performance and also the quality of up-sampled tasks.
Fine-Tuning Graph Neural Networks via Graph Topology induced Optimal Transport
Mar 20, 2022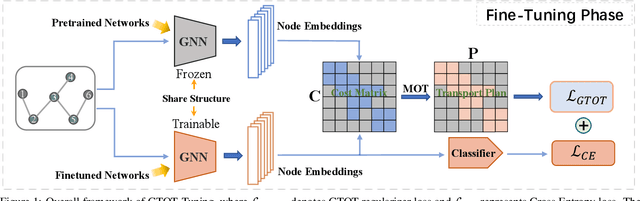
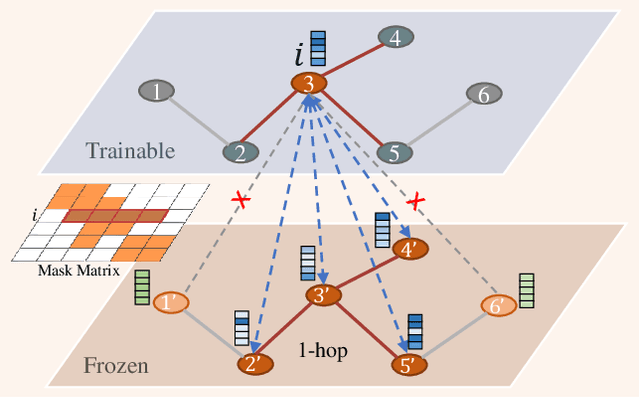

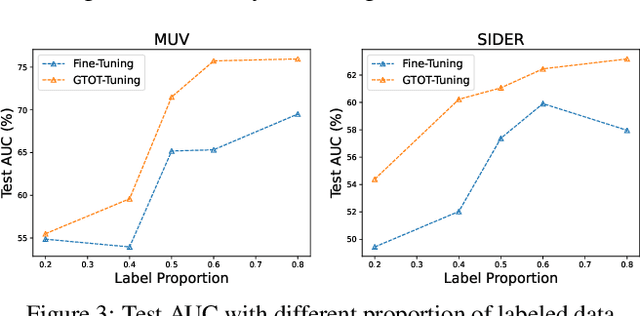
Abstract:Recently, the pretrain-finetuning paradigm has attracted tons of attention in graph learning community due to its power of alleviating the lack of labels problem in many real-world applications. Current studies use existing techniques, such as weight constraint, representation constraint, which are derived from images or text data, to transfer the invariant knowledge from the pre-train stage to fine-tuning stage. However, these methods failed to preserve invariances from graph structure and Graph Neural Network (GNN) style models. In this paper, we present a novel optimal transport-based fine-tuning framework called GTOT-Tuning, namely, Graph Topology induced Optimal Transport fine-Tuning, for GNN style backbones. GTOT-Tuning is required to utilize the property of graph data to enhance the preservation of representation produced by fine-tuned networks. Toward this goal, we formulate graph local knowledge transfer as an Optimal Transport (OT) problem with a structural prior and construct the GTOT regularizer to constrain the fine-tuned model behaviors. By using the adjacency relationship amongst nodes, the GTOT regularizer achieves node-level optimal transport procedures and reduces redundant transport procedures, resulting in efficient knowledge transfer from the pre-trained models. We evaluate GTOT-Tuning on eight downstream tasks with various GNN backbones and demonstrate that it achieves state-of-the-art fine-tuning performance for GNNs.
DrugOOD: Out-of-Distribution (OOD) Dataset Curator and Benchmark for AI-aided Drug Discovery -- A Focus on Affinity Prediction Problems with Noise Annotations
Jan 24, 2022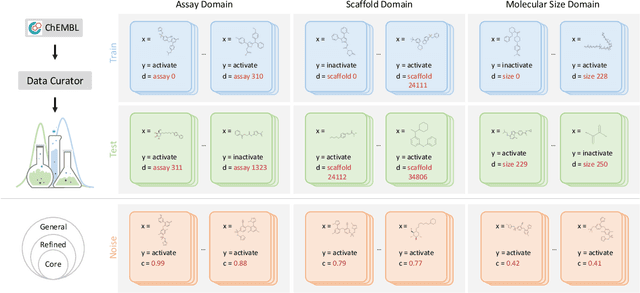
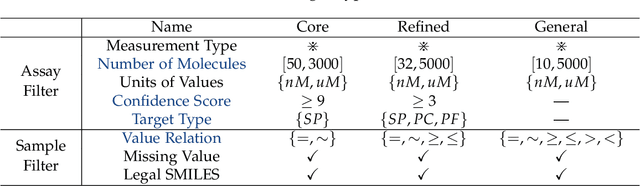
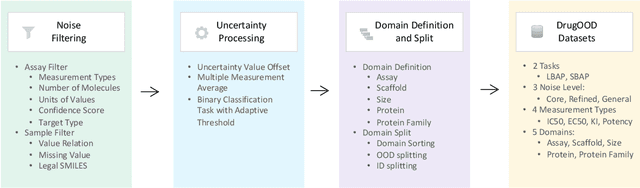
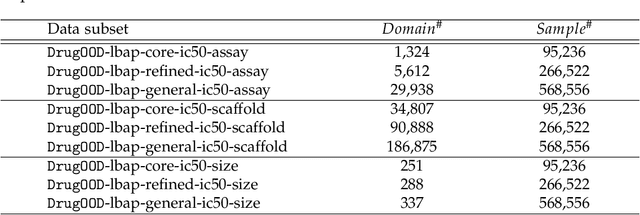
Abstract:AI-aided drug discovery (AIDD) is gaining increasing popularity due to its promise of making the search for new pharmaceuticals quicker, cheaper and more efficient. In spite of its extensive use in many fields, such as ADMET prediction, virtual screening, protein folding and generative chemistry, little has been explored in terms of the out-of-distribution (OOD) learning problem with \emph{noise}, which is inevitable in real world AIDD applications. In this work, we present DrugOOD, a systematic OOD dataset curator and benchmark for AI-aided drug discovery, which comes with an open-source Python package that fully automates the data curation and OOD benchmarking processes. We focus on one of the most crucial problems in AIDD: drug target binding affinity prediction, which involves both macromolecule (protein target) and small-molecule (drug compound). In contrast to only providing fixed datasets, DrugOOD offers automated dataset curator with user-friendly customization scripts, rich domain annotations aligned with biochemistry knowledge, realistic noise annotations and rigorous benchmarking of state-of-the-art OOD algorithms. Since the molecular data is often modeled as irregular graphs using graph neural network (GNN) backbones, DrugOOD also serves as a valuable testbed for \emph{graph OOD learning} problems. Extensive empirical studies have shown a significant performance gap between in-distribution and out-of-distribution experiments, which highlights the need to develop better schemes that can allow for OOD generalization under noise for AIDD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge