Yikun Zhang
University of Washington, Seattle
Transfer Learning Through Conditional Quantile Matching
Feb 02, 2026Abstract:We introduce a transfer learning framework for regression that leverages heterogeneous source domains to improve predictive performance in a data-scarce target domain. Our approach learns a conditional generative model separately for each source domain and calibrates the generated responses to the target domain via conditional quantile matching. This distributional alignment step corrects general discrepancies between source and target domains without imposing restrictive assumptions such as covariate or label shift. The resulting framework provides a principled and flexible approach to high-quality data augmentation for downstream learning tasks in the target domain. From a theoretical perspective, we show that an empirical risk minimizer (ERM) trained on the augmented dataset achieves a tighter excess risk bound than the target-only ERM under mild conditions. In particular, we establish new convergence rates for the quantile matching estimator that governs the transfer bias-variance tradeoff. From a practical perspective, extensive simulations and real data applications demonstrate that the proposed method consistently improves prediction accuracy over target-only learning and competing transfer learning methods.
Iterative Diffusion-Refined Neural Attenuation Fields for Multi-Source Stationary CT Reconstruction: NAF Meets Diffusion Model
Nov 18, 2025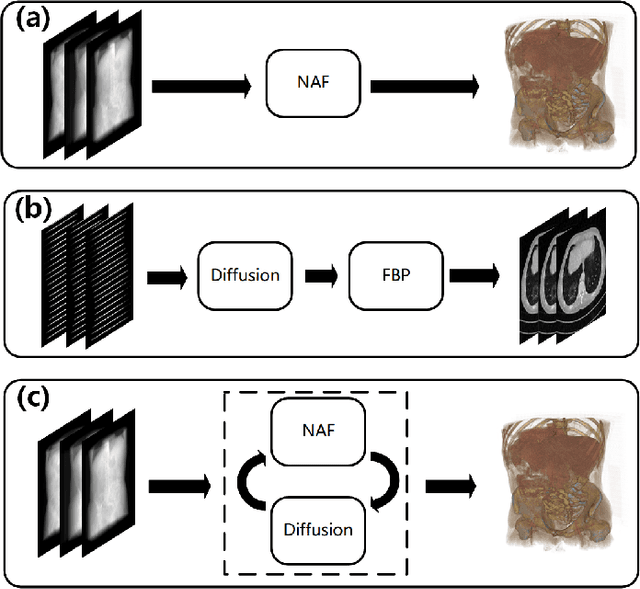
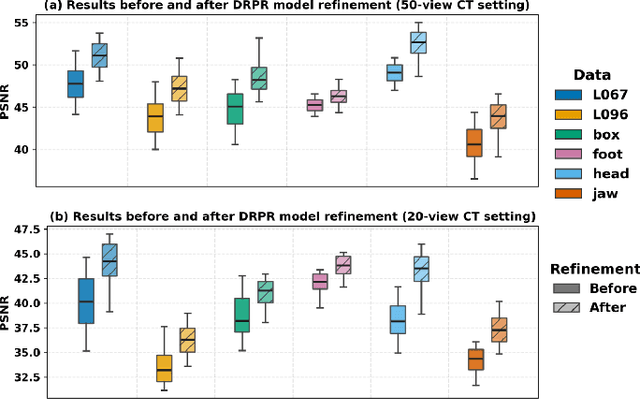
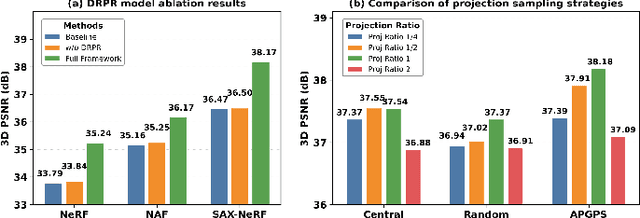
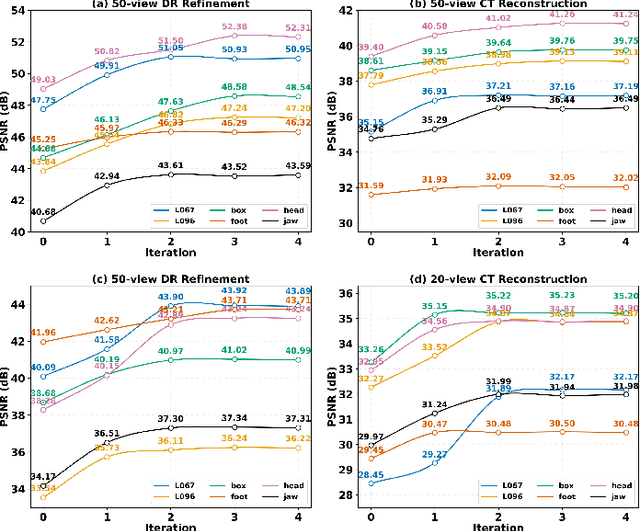
Abstract:Multi-source stationary computed tomography (CT) has recently attracted attention for its ability to achieve rapid image reconstruction, making it suitable for time-sensitive clinical and industrial applications. However, practical systems are often constrained by ultra-sparse-view sampling, which significantly degrades reconstruction quality. Traditional methods struggle under ultra-sparse-view settings, where interpolation becomes inaccurate and the resulting reconstructions are unsatisfactory. To address this challenge, this study proposes Diffusion-Refined Neural Attenuation Fields (Diff-NAF), an iterative framework tailored for multi-source stationary CT under ultra-sparse-view conditions. Diff-NAF combines a Neural Attenuation Field representation with a dual-branch conditional diffusion model. The process begins by training an initial NAF using ultra-sparse-view projections. New projections are then generated through an Angle-Prior Guided Projection Synthesis strategy that exploits inter view priors, and are subsequently refined by a Diffusion-driven Reuse Projection Refinement Module. The refined projections are incorporated as pseudo-labels into the training set for the next iteration. Through iterative refinement, Diff-NAF progressively enhances projection completeness and reconstruction fidelity under ultra-sparse-view conditions, ultimately yielding high-quality CT reconstructions. Experimental results on multiple simulated 3D CT volumes and real projection data demonstrate that Diff-NAF achieves the best performance under ultra-sparse-view conditions.
A Comparative Review of RNA Language Models
May 14, 2025
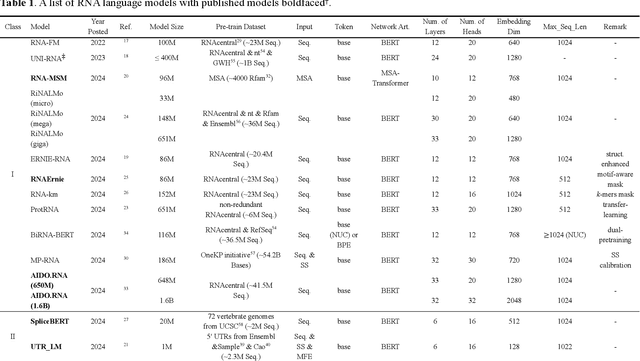

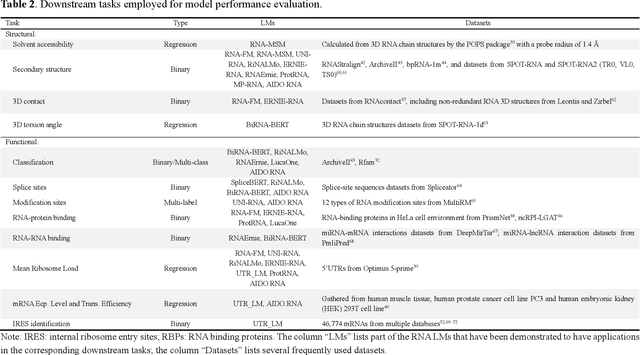
Abstract:Given usefulness of protein language models (LMs) in structure and functional inference, RNA LMs have received increased attentions in the last few years. However, these RNA models are often not compared against the same standard. Here, we divided RNA LMs into three classes (pretrained on multiple RNA types (especially noncoding RNAs), specific-purpose RNAs, and LMs that unify RNA with DNA or proteins or both) and compared 13 RNA LMs along with 3 DNA and 1 protein LMs as controls in zero-shot prediction of RNA secondary structure and functional classification. Results shows that the models doing well on secondary structure prediction often perform worse in function classification or vice versa, suggesting that more balanced unsupervised training is needed.
Doubly Robust Inference on Causal Derivative Effects for Continuous Treatments
Jan 12, 2025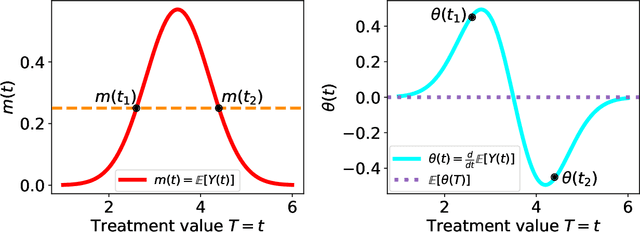
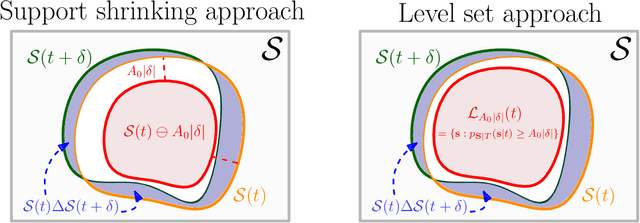
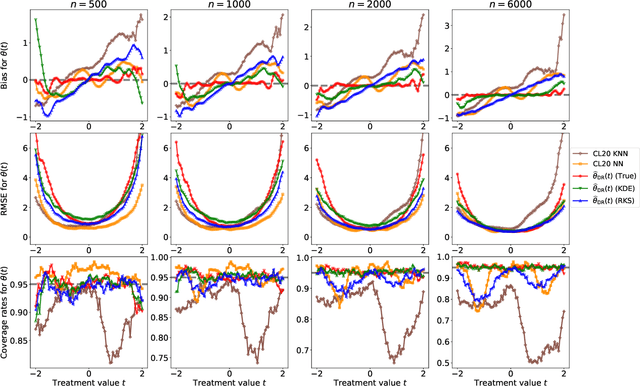
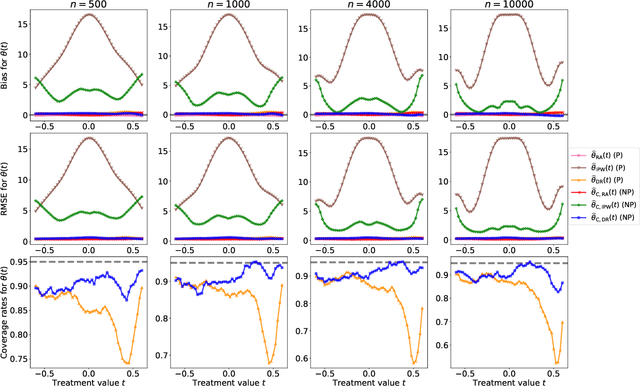
Abstract:Statistical methods for causal inference with continuous treatments mainly focus on estimating the mean potential outcome function, commonly known as the dose-response curve. However, it is often not the dose-response curve but its derivative function that signals the treatment effect. In this paper, we investigate nonparametric inference on the derivative of the dose-response curve with and without the positivity condition. Under the positivity and other regularity conditions, we propose a doubly robust (DR) inference method for estimating the derivative of the dose-response curve using kernel smoothing. When the positivity condition is violated, we demonstrate the inconsistency of conventional inverse probability weighting (IPW) and DR estimators, and introduce novel bias-corrected IPW and DR estimators. In all settings, our DR estimator achieves asymptotic normality at the standard nonparametric rate of convergence. Additionally, our approach reveals an interesting connection to nonparametric support and level set estimation problems. Finally, we demonstrate the applicability of our proposed estimators through simulations and a case study of evaluating a job training program.
BLADE: Benchmarking Language Model Agents for Data-Driven Science
Aug 20, 2024Abstract:Data-driven scientific discovery requires the iterative integration of scientific domain knowledge, statistical expertise, and an understanding of data semantics to make nuanced analytical decisions, e.g., about which variables, transformations, and statistical models to consider. LM-based agents equipped with planning, memory, and code execution capabilities have the potential to support data-driven science. However, evaluating agents on such open-ended tasks is challenging due to multiple valid approaches, partially correct steps, and different ways to express the same decisions. To address these challenges, we present BLADE, a benchmark to automatically evaluate agents' multifaceted approaches to open-ended research questions. BLADE consists of 12 datasets and research questions drawn from existing scientific literature, with ground truth collected from independent analyses by expert data scientists and researchers. To automatically evaluate agent responses, we developed corresponding computational methods to match different representations of analyses to this ground truth. Though language models possess considerable world knowledge, our evaluation shows that they are often limited to basic analyses. However, agents capable of interacting with the underlying data demonstrate improved, but still non-optimal, diversity in their analytical decision making. Our work enables the evaluation of agents for data-driven science and provides researchers deeper insights into agents' analysis approaches.
Atomas: Hierarchical Alignment on Molecule-Text for Unified Molecule Understanding and Generation
Apr 23, 2024Abstract:Molecule-and-text cross-modal representation learning has emerged as a promising direction for enhancing the quality of molecular representation, thereby improving performance in various scientific fields, including drug discovery and materials science. Existing studies adopt a global alignment approach to learn the knowledge from different modalities. These global alignment approaches fail to capture fine-grained information, such as molecular fragments and their corresponding textual description, which is crucial for downstream tasks. Furthermore, it is incapable to model such information using a similar global alignment strategy due to data scarcity of paired local part annotated data from existing datasets. In this paper, we propose Atomas, a multi-modal molecular representation learning framework to jointly learn representations from SMILES string and text. We design a Hierarchical Adaptive Alignment model to concurrently learn the fine-grained fragment correspondence between two modalities and align these representations of fragments in three levels. Additionally, Atomas's end-to-end training framework incorporates the tasks of understanding and generating molecule, thereby supporting a wider range of downstream tasks. In the retrieval task, Atomas exhibits robust generalization ability and outperforms the baseline by 30.8% of recall@1 on average. In the generation task, Atomas achieves state-of-the-art results in both molecule captioning task and molecule generation task. Moreover, the visualization of the Hierarchical Adaptive Alignment model further confirms the chemical significance of our approach. Our codes can be found at https://anonymous.4open.science/r/Atomas-03C3.
Functional Protein Design with Local Domain Alignment
Apr 18, 2024



Abstract:The core challenge of de novo protein design lies in creating proteins with specific functions or properties, guided by certain conditions. Current models explore to generate protein using structural and evolutionary guidance, which only provide indirect conditions concerning functions and properties. However, textual annotations of proteins, especially the annotations for protein domains, which directly describe the protein's high-level functionalities, properties, and their correlation with target amino acid sequences, remain unexplored in the context of protein design tasks. In this paper, we propose Protein-Annotation Alignment Generation (PAAG), a multi-modality protein design framework that integrates the textual annotations extracted from protein database for controllable generation in sequence space. Specifically, within a multi-level alignment module, PAAG can explicitly generate proteins containing specific domains conditioned on the corresponding domain annotations, and can even design novel proteins with flexible combinations of different kinds of annotations. Our experimental results underscore the superiority of the aligned protein representations from PAAG over 7 prediction tasks. Furthermore, PAAG demonstrates a nearly sixfold increase in generation success rate (24.7% vs 4.7% in zinc finger, and 54.3% vs 8.7% in the immunoglobulin domain) in comparison to the existing model.
Mode and Ridge Estimation in Euclidean and Directional Product Spaces: A Mean Shift Approach
Oct 16, 2021
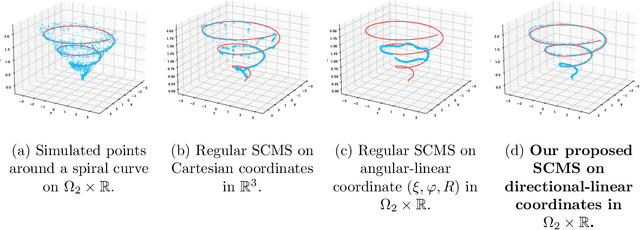
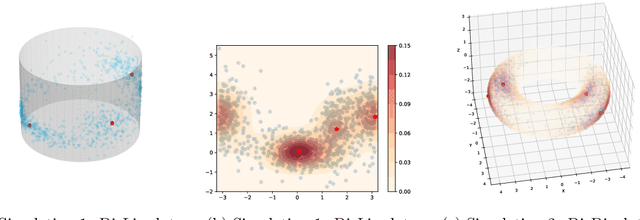

Abstract:The set of local modes and the ridge lines estimated from a dataset are important summary characteristics of the data-generating distribution. In this work, we consider estimating the local modes and ridges from point cloud data in a product space with two or more Euclidean/directional metric spaces. Specifically, we generalize the well-known (subspace constrained) mean shift algorithm to the product space setting and illuminate some pitfalls in such generalization. We derive the algorithmic convergence of the proposed method, provide practical guidelines on the implementation, and demonstrate its effectiveness on both simulated and real datasets.
Linear Convergence of the Subspace Constrained Mean Shift Algorithm: From Euclidean to Directional Data
Apr 29, 2021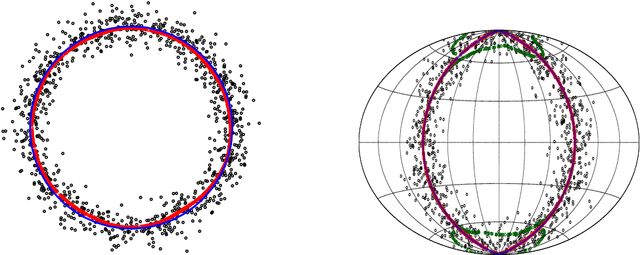
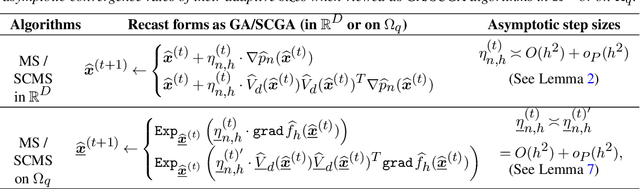
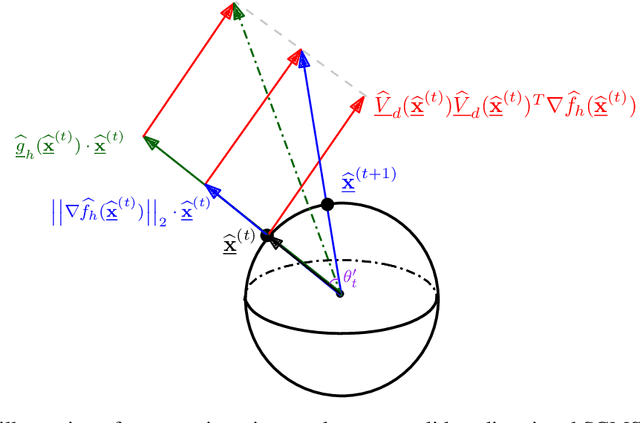
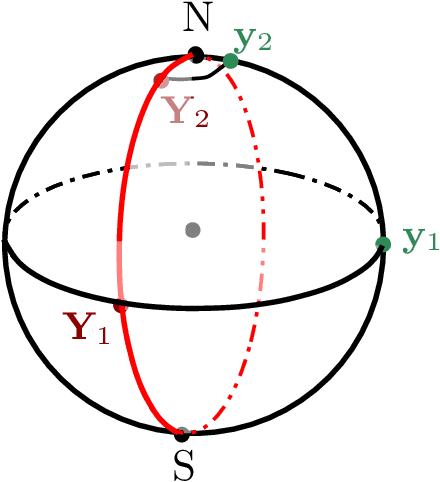
Abstract:This paper studies linear convergence of the subspace constrained mean shift (SCMS) algorithm, a well-known algorithm for identifying a density ridge defined by a kernel density estimator. By arguing that the SCMS algorithm is a special variant of a subspace constrained gradient ascent (SCGA) algorithm with an adaptive step size, we derive linear convergence of such SCGA algorithm. While the existing research focuses mainly on density ridges in the Euclidean space, we generalize density ridges and the SCMS algorithm to directional data. In particular, we establish the stability theorem of density ridges with directional data and prove the linear convergence of our proposed directional SCMS algorithm.
The EM Perspective of Directional Mean Shift Algorithm
Jan 25, 2021
Abstract:The directional mean shift (DMS) algorithm is a nonparametric method for pursuing local modes of densities defined by kernel density estimators on the unit hypersphere. In this paper, we show that any DMS iteration can be viewed as a generalized Expectation-Maximization (EM) algorithm; in particular, when the von Mises kernel is applied, it becomes an exact EM algorithm. Under the (generalized) EM framework, we provide a new proof for the ascending property of density estimates and demonstrate the global convergence of directional mean shift sequences. Finally, we give a new insight into the linear convergence of the DMS algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge