Chaohao Yuan
ParaFormer: A Generalized PageRank Graph Transformer for Graph Representation Learning
Dec 16, 2025Abstract:Graph Transformers (GTs) have emerged as a promising graph learning tool, leveraging their all-pair connected property to effectively capture global information. To address the over-smoothing problem in deep GNNs, global attention was initially introduced, eliminating the necessity for using deep GNNs. However, through empirical and theoretical analysis, we verify that the introduced global attention exhibits severe over-smoothing, causing node representations to become indistinguishable due to its inherent low-pass filtering. This effect is even stronger than that observed in GNNs. To mitigate this, we propose PageRank Transformer (ParaFormer), which features a PageRank-enhanced attention module designed to mimic the behavior of deep Transformers. We theoretically and empirically demonstrate that ParaFormer mitigates over-smoothing by functioning as an adaptive-pass filter. Experiments show that ParaFormer achieves consistent performance improvements across both node classification and graph classification tasks on 11 datasets ranging from thousands to millions of nodes, validating its efficacy. The supplementary material, including code and appendix, can be found in https://github.com/chaohaoyuan/ParaFormer.
ASD Classification on Dynamic Brain Connectome using Temporal Random Walk with Transformer-based Dynamic Network Embedding
Mar 16, 2025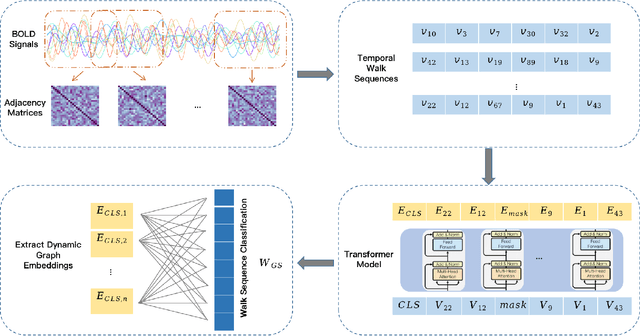
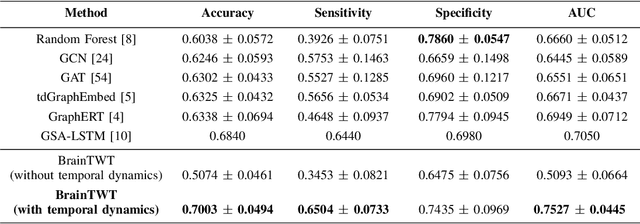

Abstract:Autism Spectrum Disorder (ASD) is a complex neurological condition characterized by varied developmental impairments, especially in communication and social interaction. Accurate and early diagnosis of ASD is crucial for effective intervention, which is enhanced by richer representations of brain activity. The brain functional connectome, which refers to the statistical relationships between different brain regions measured through neuroimaging, provides crucial insights into brain function. Traditional static methods often fail to capture the dynamic nature of brain activity, in contrast, dynamic brain connectome analysis provides a more comprehensive view by capturing the temporal variations in the brain. We propose BrainTWT, a novel dynamic network embedding approach that captures temporal evolution of the brain connectivity over time and considers also the dynamics between different temporal network snapshots. BrainTWT employs temporal random walks to capture dynamics across different temporal network snapshots and leverages the Transformer's ability to model long term dependencies in sequential data to learn the discriminative embeddings from these temporal sequences using temporal structure prediction tasks. The experimental evaluation, utilizing the Autism Brain Imaging Data Exchange (ABIDE) dataset, demonstrates that BrainTWT outperforms baseline methods in ASD classification.
A Survey of Graph Transformers: Architectures, Theories and Applications
Feb 23, 2025



Abstract:Graph Transformers (GTs) have demonstrated a strong capability in modeling graph structures by addressing the intrinsic limitations of graph neural networks (GNNs), such as over-smoothing and over-squashing. Recent studies have proposed diverse architectures, enhanced explainability, and practical applications for Graph Transformers. In light of these rapid developments, we conduct a comprehensive review of Graph Transformers, covering aspects such as their architectures, theoretical foundations, and applications within this survey. We categorize the architecture of Graph Transformers according to their strategies for processing structural information, including graph tokenization, positional encoding, structure-aware attention and model ensemble. Furthermore, from the theoretical perspective, we examine the expressivity of Graph Transformers in various discussed architectures and contrast them with other advanced graph learning algorithms to discover the connections. Furthermore, we provide a summary of the practical applications where Graph Transformers have been utilized, such as molecule, protein, language, vision traffic, brain and material data. At the end of this survey, we will discuss the current challenges and prospective directions in Graph Transformers for potential future research.
Atomas: Hierarchical Alignment on Molecule-Text for Unified Molecule Understanding and Generation
Apr 23, 2024Abstract:Molecule-and-text cross-modal representation learning has emerged as a promising direction for enhancing the quality of molecular representation, thereby improving performance in various scientific fields, including drug discovery and materials science. Existing studies adopt a global alignment approach to learn the knowledge from different modalities. These global alignment approaches fail to capture fine-grained information, such as molecular fragments and their corresponding textual description, which is crucial for downstream tasks. Furthermore, it is incapable to model such information using a similar global alignment strategy due to data scarcity of paired local part annotated data from existing datasets. In this paper, we propose Atomas, a multi-modal molecular representation learning framework to jointly learn representations from SMILES string and text. We design a Hierarchical Adaptive Alignment model to concurrently learn the fine-grained fragment correspondence between two modalities and align these representations of fragments in three levels. Additionally, Atomas's end-to-end training framework incorporates the tasks of understanding and generating molecule, thereby supporting a wider range of downstream tasks. In the retrieval task, Atomas exhibits robust generalization ability and outperforms the baseline by 30.8% of recall@1 on average. In the generation task, Atomas achieves state-of-the-art results in both molecule captioning task and molecule generation task. Moreover, the visualization of the Hierarchical Adaptive Alignment model further confirms the chemical significance of our approach. Our codes can be found at https://anonymous.4open.science/r/Atomas-03C3.
Functional Protein Design with Local Domain Alignment
Apr 18, 2024



Abstract:The core challenge of de novo protein design lies in creating proteins with specific functions or properties, guided by certain conditions. Current models explore to generate protein using structural and evolutionary guidance, which only provide indirect conditions concerning functions and properties. However, textual annotations of proteins, especially the annotations for protein domains, which directly describe the protein's high-level functionalities, properties, and their correlation with target amino acid sequences, remain unexplored in the context of protein design tasks. In this paper, we propose Protein-Annotation Alignment Generation (PAAG), a multi-modality protein design framework that integrates the textual annotations extracted from protein database for controllable generation in sequence space. Specifically, within a multi-level alignment module, PAAG can explicitly generate proteins containing specific domains conditioned on the corresponding domain annotations, and can even design novel proteins with flexible combinations of different kinds of annotations. Our experimental results underscore the superiority of the aligned protein representations from PAAG over 7 prediction tasks. Furthermore, PAAG demonstrates a nearly sixfold increase in generation success rate (24.7% vs 4.7% in zinc finger, and 54.3% vs 8.7% in the immunoglobulin domain) in comparison to the existing model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge