Hieu Pham
Brain Network Analysis Based on Fine-tuned Self-supervised Model for Brain Disease Diagnosis
Jun 13, 2025Abstract:Functional brain network analysis has become an indispensable tool for brain disease analysis. It is profoundly impacted by deep learning methods, which can characterize complex connections between ROIs. However, the research on foundation models of brain network is limited and constrained to a single dimension, which restricts their extensive application in neuroscience. In this study, we propose a fine-tuned brain network model for brain disease diagnosis. It expands brain region representations across multiple dimensions based on the original brain network model, thereby enhancing its generalizability. Our model consists of two key modules: (1)an adapter module that expands brain region features across different dimensions. (2)a fine-tuned foundation brain network model, based on self-supervised learning and pre-trained on fMRI data from thousands of participants. Specifically, its transformer block is able to effectively extract brain region features and compute the inter-region associations. Moreover, we derive a compact latent representation of the brain network for brain disease diagnosis. Our downstream experiments in this study demonstrate that the proposed model achieves superior performance in brain disease diagnosis, which potentially offers a promising approach in brain network analysis research.
Semise: Semi-supervised learning for severity representation in medical image
Jan 07, 2025Abstract:This paper introduces SEMISE, a novel method for representation learning in medical imaging that combines self-supervised and supervised learning. By leveraging both labeled and augmented data, SEMISE addresses the challenge of data scarcity and enhances the encoder's ability to extract meaningful features. This integrated approach leads to more informative representations, improving performance on downstream tasks. As result, our approach achieved a 12% improvement in classification and a 3% improvement in segmentation, outperforming existing methods. These results demonstrate the potential of SIMESE to advance medical image analysis and offer more accurate solutions for healthcare applications, particularly in contexts where labeled data is limited.
Quantitative Gait Analysis from Single RGB Videos Using a Dual-Input Transformer-Based Network
Jan 03, 2025Abstract:Gait and movement analysis have become a well-established clinical tool for diagnosing health conditions, monitoring disease progression for a wide spectrum of diseases, and to implement and assess treatment, surgery and or rehabilitation interventions. However, quantitative motion assessment remains limited to costly motion capture systems and specialized personnel, restricting its accessibility and broader application. Recent advancements in deep neural networks have enabled quantitative movement analysis using single-camera videos, offering an accessible alternative to conventional motion capture systems. In this paper, we present an efficient approach for clinical gait analysis through a dual-pattern input convolutional Transformer network. The proposed system leverages a dual-input Transformer model to estimate essential gait parameters from single RGB videos captured by a single-view camera. The system demonstrates high accuracy in estimating critical metrics such as the gait deviation index (GDI), knee flexion angle, step length, and walking cadence, validated on a dataset of individuals with movement disorders. Notably, our approach surpasses state-of-the-art methods in various scenarios, using fewer resources and proving highly suitable for clinical application, particularly in resource-constrained environments.
D-SarcNet: A Dual-stream Deep Learning Framework for Automatic Analysis of Sarcomere Structures in Fluorescently Labeled hiPSC-CMs
Oct 19, 2024



Abstract:Human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) are a powerful tool in advancing cardiovascular research and clinical applications. The maturation of sarcomere organization in hiPSC-CMs is crucial, as it supports the contractile function and structural integrity of these cells. Traditional methods for assessing this maturation like manual annotation and feature extraction are labor-intensive, time-consuming, and unsuitable for high-throughput analysis. To address this, we propose D-SarcNet, a dual-stream deep learning framework that takes fluorescent hiPSC-CM single-cell images as input and outputs the stage of the sarcomere structural organization on a scale from 1.0 to 5.0. The framework also integrates Fast Fourier Transform (FFT), deep learning-generated local patterns, and gradient magnitude to capture detailed structural information at both global and local levels. Experiments on a publicly available dataset from the Allen Institute for Cell Science show that the proposed approach not only achieves a Spearman correlation of 0.868 marking a 3.7% improvement over the previous state-of-the-art but also significantly enhances other key performance metrics, including MSE, MAE, and R2 score. Beyond establishing a new state-of-the-art in sarcomere structure assessment from hiPSC-CM images, our ablation studies highlight the significance of integrating global and local information to enhance deep learning networks ability to discern and learn vital visual features of sarcomere structure.
SarcNet: A Novel AI-based Framework to Automatically Analyze and Score Sarcomere Organizations in Fluorescently Tagged hiPSC-CMs
May 28, 2024Abstract:Quantifying sarcomere structure organization in human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) is crucial for understanding cardiac disease pathology, improving drug screening, and advancing regenerative medicine. Traditional methods, such as manual annotation and Fourier transform analysis, are labor-intensive, error-prone, and lack high-throughput capabilities. In this study, we present a novel deep learning-based framework that leverages cell images and integrates cell features to automatically evaluate the sarcomere structure of hiPSC-CMs from the onset of differentiation. This framework overcomes the limitations of traditional methods through automated, high-throughput analysis, providing consistent, reliable results while accurately detecting complex sarcomere patterns across diverse samples. The proposed framework contains the SarcNet, a linear layers-added ResNet-18 module, to output a continuous score ranging from one to five that captures the level of sarcomere structure organization. It is trained and validated on an open-source dataset of hiPSC-CMs images with the endogenously GFP-tagged alpha-actinin-2 structure developed by the Allen Institute for Cell Science (AICS). SarcNet achieves a Spearman correlation of 0.831 with expert evaluations, demonstrating superior performance and an improvement of 0.075 over the current state-of-the-art approach, which uses Linear Regression. Our results also show a consistent pattern of increasing organization from day 18 to day 32 of differentiation, aligning with expert evaluations. By integrating the quantitative features calculated directly from the images with the visual features learned during the deep learning model, our framework offers a more comprehensive and accurate assessment, thereby enhancing the further utility of hiPSC-CMs in medical research and therapy development.
ConPro: Learning Severity Representation for Medical Images using Contrastive Learning and Preference Optimization
Apr 29, 2024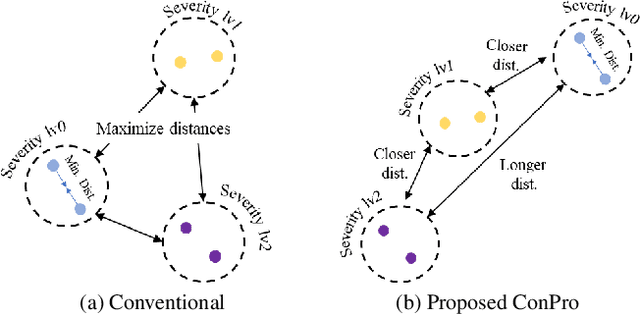
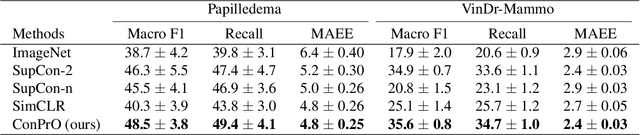
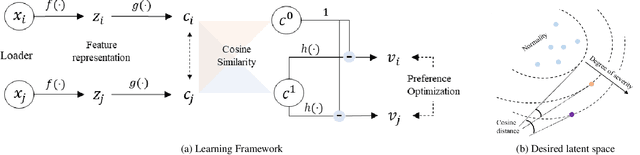
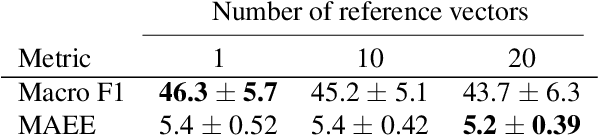
Abstract:Understanding the severity of conditions shown in images in medical diagnosis is crucial, serving as a key guide for clinical assessment, treatment, as well as evaluating longitudinal progression. This paper proposes Con- PrO: a novel representation learning method for severity assessment in medical images using Contrastive learningintegrated Preference Optimization. Different from conventional contrastive learning methods that maximize the distance between classes, ConPrO injects into the latent vector the distance preference knowledge between various severity classes and the normal class. We systematically examine the key components of our framework to illuminate how contrastive prediction tasks acquire valuable representations. We show that our representation learning framework offers valuable severity ordering in the feature space while outperforming previous state-of-the-art methods on classification tasks. We achieve a 6% and 20% relative improvement compared to a supervised and a self-supervised baseline, respectively. In addition, we derived discussions on severity indicators and related applications of preference comparison in the medical domain.
DoReMi: Optimizing Data Mixtures Speeds Up Language Model Pretraining
May 24, 2023Abstract:The mixture proportions of pretraining data domains (e.g., Wikipedia, books, web text) greatly affect language model (LM) performance. In this paper, we propose Domain Reweighting with Minimax Optimization (DoReMi), which first trains a small proxy model using group distributionally robust optimization (Group DRO) over domains to produce domain weights (mixture proportions) without knowledge of downstream tasks. We then resample a dataset with these domain weights and train a larger, full-sized model. In our experiments, we use DoReMi on a 280M-parameter proxy model to find domain weights for training an 8B-parameter model (30x larger) more efficiently. On The Pile, DoReMi improves perplexity across all domains, even when it downweights a domain. DoReMi improves average few-shot downstream accuracy by 6.5% points over a baseline model trained using The Pile's default domain weights and reaches the baseline accuracy with 2.6x fewer training steps. On the GLaM dataset, DoReMi, which has no knowledge of downstream tasks, even matches the performance of using domain weights tuned on downstream tasks.
Multimodal contrastive learning for diagnosing cardiovascular diseases from electrocardiography (ECG) signals and patient metadata
Apr 18, 2023Abstract:This work discusses the use of contrastive learning and deep learning for diagnosing cardiovascular diseases from electrocardiography (ECG) signals. While the ECG signals usually contain 12 leads (channels), many healthcare facilities and devices lack access to all these 12 leads. This raises the problem of how to use only fewer ECG leads to produce meaningful diagnoses with high performance. We introduce a simple experiment to test whether contrastive learning can be applied to this task. More specifically, we added the similarity between the embedding vectors when the 12 leads signal and the fewer leads ECG signal to the loss function to bring these representations closer together. Despite its simplicity, this has been shown to have improved the performance of diagnosing with all lead combinations, proving the potential of contrastive learning on this task.
Ensemble Learning of Myocardial Displacements for Myocardial Infarction Detection in Echocardiography
Mar 12, 2023



Abstract:Early detection and localization of myocardial infarction (MI) can reduce the severity of cardiac damage through timely treatment interventions. In recent years, deep learning techniques have shown promise for detecting MI in echocardiographic images. However, there has been no examination of how segmentation accuracy affects MI classification performance and the potential benefits of using ensemble learning approaches. Our study investigates this relationship and introduces a robust method that combines features from multiple segmentation models to improve MI classification performance by leveraging ensemble learning. Our method combines myocardial segment displacement features from multiple segmentation models, which are then input into a typical classifier to estimate the risk of MI. We validated the proposed approach on two datasets: the public HMC-QU dataset (109 echocardiograms) for training and validation, and an E-Hospital dataset (60 echocardiograms) from a local clinical site in Vietnam for independent testing. Model performance was evaluated based on accuracy, sensitivity, and specificity. The proposed approach demonstrated excellent performance in detecting MI. The results showed that the proposed approach outperformed the state-of-the-art feature-based method. Further research is necessary to determine its potential use in clinical settings as a tool to assist cardiologists and technicians with objective assessments and reduce dependence on operator subjectivity. Our research codes are available on GitHub at https://github.com/vinuni-vishc/mi-detection-echo.
Symbolic Discovery of Optimization Algorithms
Feb 17, 2023



Abstract:We present a method to formulate algorithm discovery as program search, and apply it to discover optimization algorithms for deep neural network training. We leverage efficient search techniques to explore an infinite and sparse program space. To bridge the large generalization gap between proxy and target tasks, we also introduce program selection and simplification strategies. Our method discovers a simple and effective optimization algorithm, $\textbf{Lion}$ ($\textit{Evo$\textbf{L}$ved S$\textbf{i}$gn M$\textbf{o}$me$\textbf{n}$tum}$). It is more memory-efficient than Adam as it only keeps track of the momentum. Different from adaptive optimizers, its update has the same magnitude for each parameter calculated through the sign operation. We compare Lion with widely used optimizers, such as Adam and Adafactor, for training a variety of models on different tasks. On image classification, Lion boosts the accuracy of ViT by up to 2% on ImageNet and saves up to 5x the pre-training compute on JFT. On vision-language contrastive learning, we achieve 88.3% $\textit{zero-shot}$ and 91.1% $\textit{fine-tuning}$ accuracy on ImageNet, surpassing the previous best results by 2% and 0.1%, respectively. On diffusion models, Lion outperforms Adam by achieving a better FID score and reducing the training compute by up to 2.3x. For autoregressive, masked language modeling, and fine-tuning, Lion exhibits a similar or better performance compared to Adam. Our analysis of Lion reveals that its performance gain grows with the training batch size. It also requires a smaller learning rate than Adam due to the larger norm of the update produced by the sign function. Additionally, we examine the limitations of Lion and identify scenarios where its improvements are small or not statistically significant. The implementation of Lion is publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge