Changhong Jing
Brain Network Analysis Based on Fine-tuned Self-supervised Model for Brain Disease Diagnosis
Jun 13, 2025Abstract:Functional brain network analysis has become an indispensable tool for brain disease analysis. It is profoundly impacted by deep learning methods, which can characterize complex connections between ROIs. However, the research on foundation models of brain network is limited and constrained to a single dimension, which restricts their extensive application in neuroscience. In this study, we propose a fine-tuned brain network model for brain disease diagnosis. It expands brain region representations across multiple dimensions based on the original brain network model, thereby enhancing its generalizability. Our model consists of two key modules: (1)an adapter module that expands brain region features across different dimensions. (2)a fine-tuned foundation brain network model, based on self-supervised learning and pre-trained on fMRI data from thousands of participants. Specifically, its transformer block is able to effectively extract brain region features and compute the inter-region associations. Moreover, we derive a compact latent representation of the brain network for brain disease diagnosis. Our downstream experiments in this study demonstrate that the proposed model achieves superior performance in brain disease diagnosis, which potentially offers a promising approach in brain network analysis research.
DiffGAN-F2S: Symmetric and Efficient Denoising Diffusion GANs for Structural Connectivity Prediction from Brain fMRI
Sep 28, 2023Abstract:Mapping from functional connectivity (FC) to structural connectivity (SC) can facilitate multimodal brain network fusion and discover potential biomarkers for clinical implications. However, it is challenging to directly bridge the reliable non-linear mapping relations between SC and functional magnetic resonance imaging (fMRI). In this paper, a novel diffusision generative adversarial network-based fMRI-to-SC (DiffGAN-F2S) model is proposed to predict SC from brain fMRI in an end-to-end manner. To be specific, the proposed DiffGAN-F2S leverages denoising diffusion probabilistic models (DDPMs) and adversarial learning to efficiently generate high-fidelity SC through a few steps from fMRI. By designing the dual-channel multi-head spatial attention (DMSA) and graph convolutional modules, the symmetric graph generator first captures global relations among direct and indirect connected brain regions, then models the local brain region interactions. It can uncover the complex mapping relations between fMRI and structural connectivity. Furthermore, the spatially connected consistency loss is devised to constrain the generator to preserve global-local topological information for accurate intrinsic SC prediction. Testing on the public Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset, the proposed model can effectively generate empirical SC-preserved connectivity from four-dimensional imaging data and shows superior performance in SC prediction compared with other related models. Furthermore, the proposed model can identify the vast majority of important brain regions and connections derived from the empirical method, providing an alternative way to fuse multimodal brain networks and analyze clinical disease.
Generative artificial intelligence-enabled dynamic detection of nicotine-related circuits
Dec 13, 2022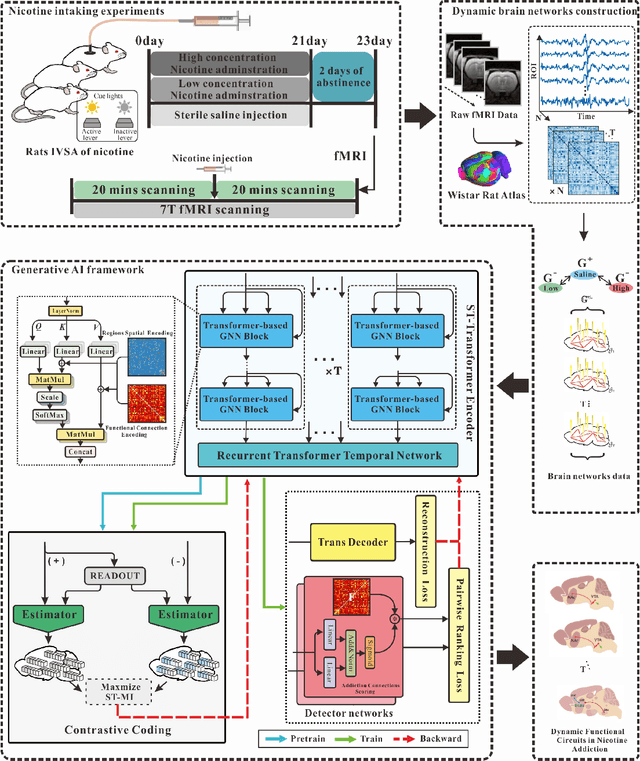
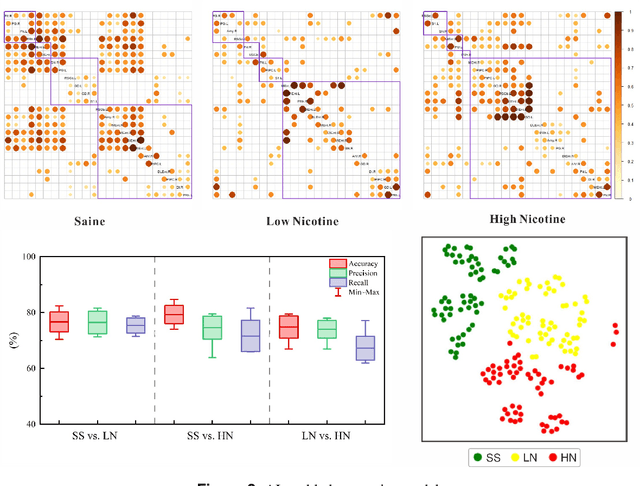

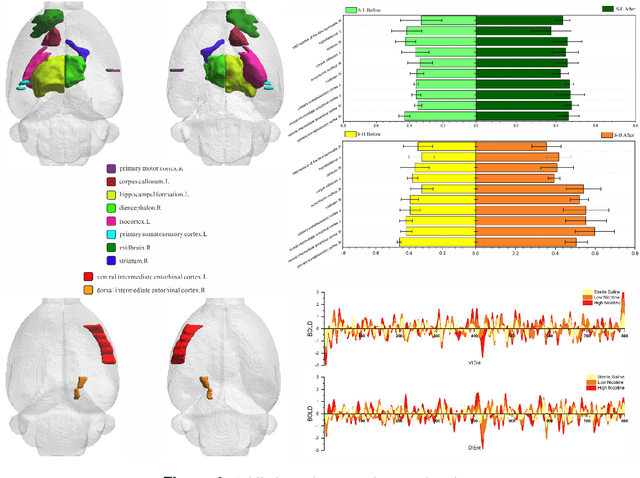
Abstract:The identification of addiction-related circuits is critical for explaining addiction processes and developing addiction treatments. And models of functional addiction circuits developed from functional imaging are an effective tool for discovering and verifying addiction circuits. However, analyzing functional imaging data of addiction and detecting functional addiction circuits still have challenges. We have developed a data-driven and end-to-end generative artificial intelligence(AI) framework to address these difficulties. The framework integrates dynamic brain network modeling and novel network architecture networks architecture, including temporal graph Transformer and contrastive learning modules. A complete workflow is formed by our generative AI framework: the functional imaging data, from neurobiological experiments, and computational modeling, to end-to-end neural networks, is transformed into dynamic nicotine addiction-related circuits. It enables the detection of addiction-related brain circuits with dynamic properties and reveals the underlying mechanisms of addiction.
Multiscale Autoencoder with Structural-Functional Attention Network for Alzheimer's Disease Prediction
Aug 09, 2022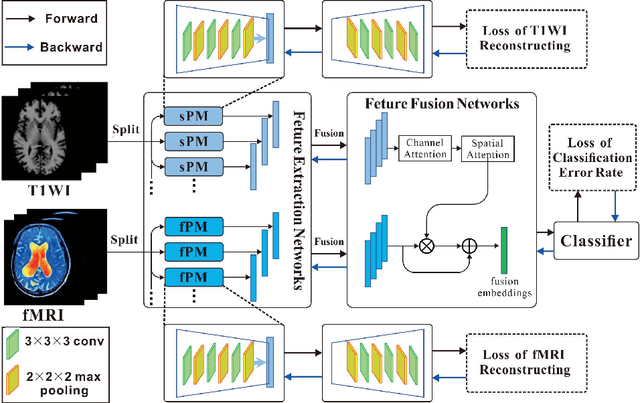

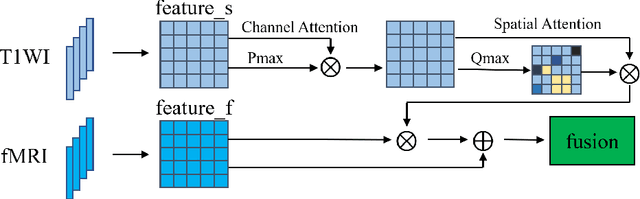
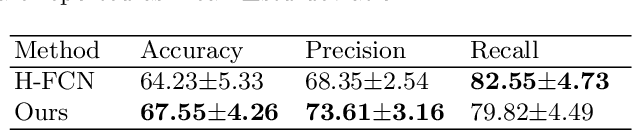
Abstract:The application of machine learning algorithms to the diagnosis and analysis of Alzheimer's disease (AD) from multimodal neuroimaging data is a current research hotspot. It remains a formidable challenge to learn brain region information and discover disease mechanisms from various magnetic resonance images (MRI). In this paper, we propose a simple but highly efficient end-to-end model, a multiscale autoencoder with structural-functional attention network (MASAN) to extract disease-related representations using T1-weighted Imaging (T1WI) and functional MRI (fMRI). Based on the attention mechanism, our model effectively learns the fused features of brain structure and function and finally is trained for the classification of Alzheimer's disease. Compared with the fully convolutional network, the proposed method has further improvement in both accuracy and precision, leading by 3% to 5%. By visualizing the extracted embedding, the empirical results show that there are higher weights on putative AD-related brain regions (such as the hippocampus, amygdala, etc.), and these regions are much more informative in anatomical studies. Conversely, the cerebellum, parietal lobe, thalamus, brain stem, and ventral diencephalon have little predictive contribution.
Feature-selected Graph Spatial Attention Network for Addictive Brain-Networks Identification
Jul 05, 2022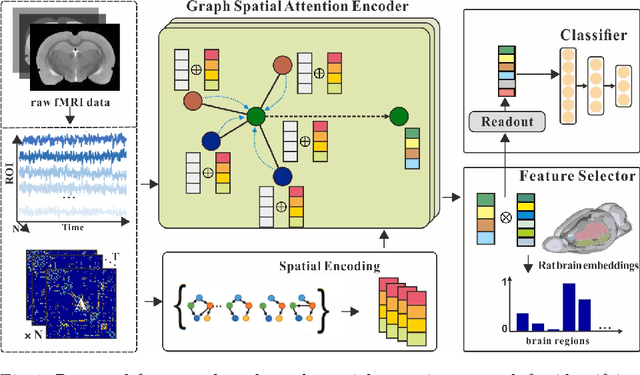

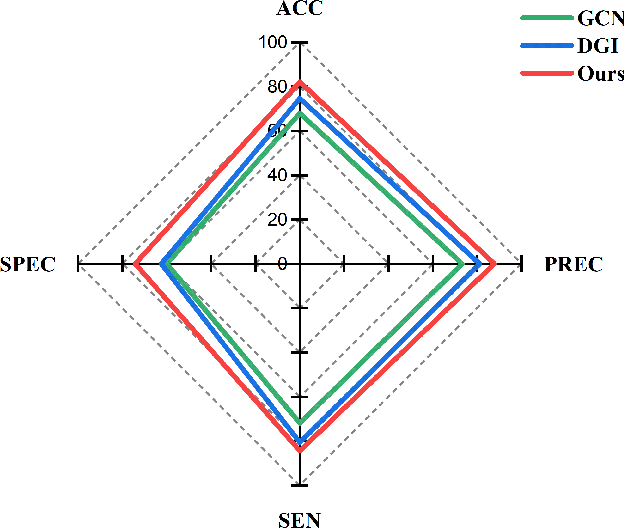
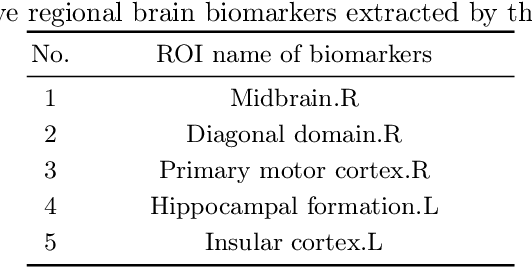
Abstract:Functional alterations in the relevant neural circuits occur from drug addiction over a certain period. And these significant alterations are also revealed by analyzing fMRI. However, because of fMRI's high dimensionality and poor signal-to-noise ratio, it is challenging to encode efficient and robust brain regional embeddings for both graph-level identification and region-level biomarkers detection tasks between nicotine addiction (NA) and healthy control (HC) groups. In this work, we represent the fMRI of the rat brain as a graph with biological attributes and propose a novel feature-selected graph spatial attention network(FGSAN) to extract the biomarkers of addiction and identify from these brain networks. Specially, a graph spatial attention encoder is employed to capture the features of spatiotemporal brain networks with spatial information. The method simultaneously adopts a Bayesian feature selection strategy to optimize the model and improve classification task by constraining features. Experiments on an addiction-related neural imaging dataset show that the proposed model can obtain superior performance and detect interpretable biomarkers associated with addiction-relevant neural circuits.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge