Qiankun Zuo
MGML: A Plug-and-Play Meta-Guided Multi-Modal Learning Framework for Incomplete Multimodal Brain Tumor Segmentation
Dec 30, 2025Abstract:Leveraging multimodal information from Magnetic Resonance Imaging (MRI) plays a vital role in lesion segmentation, especially for brain tumors. However, in clinical practice, multimodal MRI data are often incomplete, making it challenging to fully utilize the available information. Therefore, maximizing the utilization of this incomplete multimodal information presents a crucial research challenge. We present a novel meta-guided multi-modal learning (MGML) framework that comprises two components: meta-parameterized adaptive modality fusion and consistency regularization module. The meta-parameterized adaptive modality fusion (Meta-AMF) enables the model to effectively integrate information from multiple modalities under varying input conditions. By generating adaptive soft-label supervision signals based on the available modalities, Meta-AMF explicitly promotes more coherent multimodal fusion. In addition, the consistency regularization module enhances segmentation performance and implicitly reinforces the robustness and generalization of the overall framework. Notably, our approach does not alter the original model architecture and can be conveniently integrated into the training pipeline for end-to-end model optimization. We conducted extensive experiments on the public BraTS2020 and BraTS2023 datasets. Compared to multiple state-of-the-art methods from previous years, our method achieved superior performance. On BraTS2020, for the average Dice scores across fifteen missing modality combinations, building upon the baseline, our method obtained scores of 87.55, 79.36, and 62.67 for the whole tumor (WT), the tumor core (TC), and the enhancing tumor (ET), respectively. We have made our source code publicly available at https://github.com/worldlikerr/MGML.
Brain Diffuser with Hierarchical Transformer for MCI Causality Analysis
Dec 14, 2023Abstract:Effective connectivity estimation plays a crucial role in understanding the interactions and information flow between different brain regions. However, the functional time series used for estimating effective connentivity is derived from certain software, which may lead to large computing errors because of different parameter settings and degrade the ability to model complex causal relationships between brain regions. In this paper, a brain diffuser with hierarchical transformer (BDHT) is proposed to estimate effective connectivity for mild cognitive impairment (MCI) analysis. To our best knowledge, the proposed brain diffuer is the first generative model to apply diffusion models in the application of generating and analyzing multimodal brain networks. Specifically, the BDHT leverages the structural connectivity to guide the reverse processes in an efficient way. It makes the denoising process more reliable and guarantees effective connectivity estimation accuracy. To improve denoising quality, the hierarchical denoising transformer is designed to learn multi-scale features in topological space. Furthermore, the GraphConFormer block can concentrate on both global and adjacent connectivity information. By stacking the multi-head attention and graph convolutional network, the proposed model enhances structure-function complementarity and improves the ability in noise estimation. Experimental evaluations of the denoising diffusion model demonstrate its effectiveness in estimating effective connectivity. The method achieves superior performance in terms of accuracy and robustness compared to existing approaches. It can captures both unidirectal and bidirectional interactions between brain regions, providing a comprehensive understanding of the brain's information processing mechanisms.
Alzheimer's Disease Prediction via Brain Structural-Functional Deep Fusing Network
Oct 05, 2023Abstract:Fusing structural-functional images of the brain has shown great potential to analyze the deterioration of Alzheimer's disease (AD). However, it is a big challenge to effectively fuse the correlated and complementary information from multimodal neuroimages. In this paper, a novel model termed cross-modal transformer generative adversarial network (CT-GAN) is proposed to effectively fuse the functional and structural information contained in functional magnetic resonance imaging (fMRI) and diffusion tensor imaging (DTI). The CT-GAN can learn topological features and generate multimodal connectivity from multimodal imaging data in an efficient end-to-end manner. Moreover, the swapping bi-attention mechanism is designed to gradually align common features and effectively enhance the complementary features between modalities. By analyzing the generated connectivity features, the proposed model can identify AD-related brain connections. Evaluations on the public ADNI dataset show that the proposed CT-GAN can dramatically improve prediction performance and detect AD-related brain regions effectively. The proposed model also provides new insights for detecting AD-related abnormal neural circuits.
DiffGAN-F2S: Symmetric and Efficient Denoising Diffusion GANs for Structural Connectivity Prediction from Brain fMRI
Sep 28, 2023Abstract:Mapping from functional connectivity (FC) to structural connectivity (SC) can facilitate multimodal brain network fusion and discover potential biomarkers for clinical implications. However, it is challenging to directly bridge the reliable non-linear mapping relations between SC and functional magnetic resonance imaging (fMRI). In this paper, a novel diffusision generative adversarial network-based fMRI-to-SC (DiffGAN-F2S) model is proposed to predict SC from brain fMRI in an end-to-end manner. To be specific, the proposed DiffGAN-F2S leverages denoising diffusion probabilistic models (DDPMs) and adversarial learning to efficiently generate high-fidelity SC through a few steps from fMRI. By designing the dual-channel multi-head spatial attention (DMSA) and graph convolutional modules, the symmetric graph generator first captures global relations among direct and indirect connected brain regions, then models the local brain region interactions. It can uncover the complex mapping relations between fMRI and structural connectivity. Furthermore, the spatially connected consistency loss is devised to constrain the generator to preserve global-local topological information for accurate intrinsic SC prediction. Testing on the public Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset, the proposed model can effectively generate empirical SC-preserved connectivity from four-dimensional imaging data and shows superior performance in SC prediction compared with other related models. Furthermore, the proposed model can identify the vast majority of important brain regions and connections derived from the empirical method, providing an alternative way to fuse multimodal brain networks and analyze clinical disease.
Fusing Structural and Functional Connectivities using Disentangled VAE for Detecting MCI
Jun 16, 2023
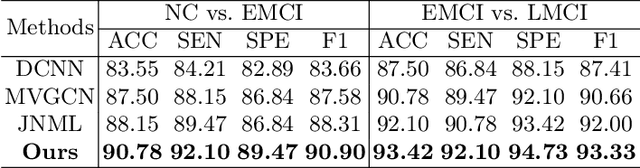
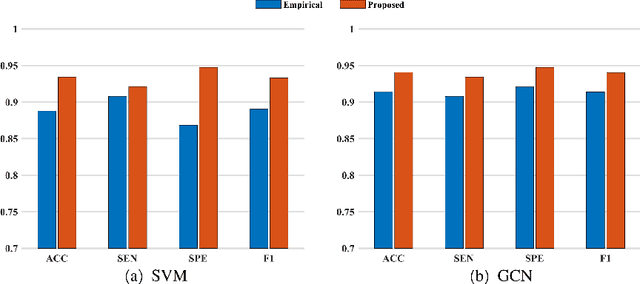
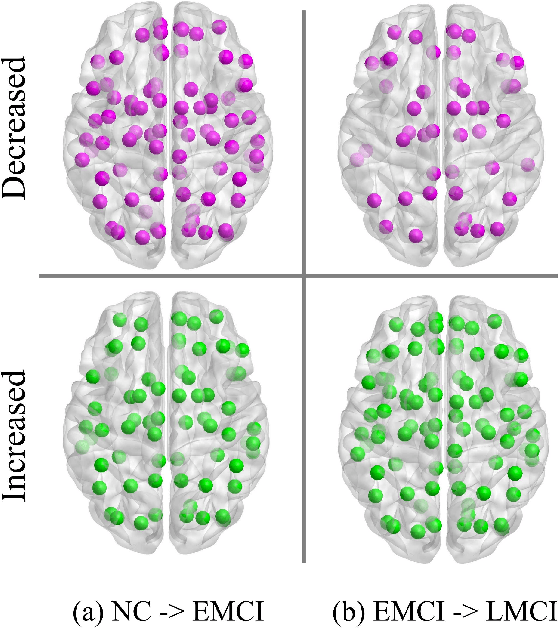
Abstract:Brain network analysis is a useful approach to studying human brain disorders because it can distinguish patients from healthy people by detecting abnormal connections. Due to the complementary information from multiple modal neuroimages, multimodal fusion technology has a lot of potential for improving prediction performance. However, effective fusion of multimodal medical images to achieve complementarity is still a challenging problem. In this paper, a novel hierarchical structural-functional connectivity fusing (HSCF) model is proposed to construct brain structural-functional connectivity matrices and predict abnormal brain connections based on functional magnetic resonance imaging (fMRI) and diffusion tensor imaging (DTI). Specifically, the prior knowledge is incorporated into the separators for disentangling each modality of information by the graph convolutional networks (GCN). And a disentangled cosine distance loss is devised to ensure the disentanglement's effectiveness. Moreover, the hierarchical representation fusion module is designed to effectively maximize the combination of relevant and effective features between modalities, which makes the generated structural-functional connectivity more robust and discriminative in the cognitive disease analysis. Results from a wide range of tests performed on the public Alzheimer's Disease Neuroimaging Initiative (ADNI) database show that the proposed model performs better than competing approaches in terms of classification evaluation. In general, the proposed HSCF model is a promising model for generating brain structural-functional connectivities and identifying abnormal brain connections as cognitive disease progresses.
Brain Structure-Function Fusing Representation Learning using Adversarial Decomposed-VAE for Analyzing MCI
May 23, 2023Abstract:Integrating the brain structural and functional connectivity features is of great significance in both exploring brain science and analyzing cognitive impairment clinically. However, it remains a challenge to effectively fuse structural and functional features in exploring the brain network. In this paper, a novel brain structure-function fusing-representation learning (BSFL) model is proposed to effectively learn fused representation from diffusion tensor imaging (DTI) and resting-state functional magnetic resonance imaging (fMRI) for mild cognitive impairment (MCI) analysis. Specifically, the decomposition-fusion framework is developed to first decompose the feature space into the union of the uniform and the unique spaces for each modality, and then adaptively fuse the decomposed features to learn MCI-related representation. Moreover, a knowledge-aware transformer module is designed to automatically capture local and global connectivity features throughout the brain. Also, a uniform-unique contrastive loss is further devised to make the decomposition more effective and enhance the complementarity of structural and functional features. The extensive experiments demonstrate that the proposed model achieves better performance than other competitive methods in predicting and analyzing MCI. More importantly, the proposed model could be a potential tool for reconstructing unified brain networks and predicting abnormal connections during the degenerative processes in MCI.
Multi-resolution Spatiotemporal Enhanced Transformer Denoising with Functional Diffusive GANs for Constructing Brain Effective Connectivity in MCI analysis
May 18, 2023



Abstract:Effective connectivity can describe the causal patterns among brain regions. These patterns have the potential to reveal the pathological mechanism and promote early diagnosis and effective drug development for cognitive disease. However, the current studies mainly focus on using empirical functional time series to calculate effective connections, which may not comprehensively capture the complex causal relationships between brain regions. In this paper, a novel Multi-resolution Spatiotemporal Enhanced Transformer Denoising (MSETD) network with an adversarially functional diffusion model is proposed to map functional magnetic resonance imaging (fMRI) into effective connectivity for mild cognitive impairment (MCI) analysis. To be specific, the denoising framework leverages a conditional diffusion process that progressively translates the noise and conditioning fMRI to effective connectivity in an end-to-end manner. To ensure reverse diffusion quality and diversity, the multi-resolution enhanced transformer generator is designed to extract local and global spatiotemporal features. Furthermore, a multi-scale diffusive transformer discriminator is devised to capture the temporal patterns at different scales for generation stability. Evaluations of the ADNI datasets demonstrate the feasibility and efficacy of the proposed model. The proposed model not only achieves superior prediction performance compared with other competing methods but also identifies MCI-related causal connections that are consistent with clinical studies.
Multiscale Autoencoder with Structural-Functional Attention Network for Alzheimer's Disease Prediction
Aug 09, 2022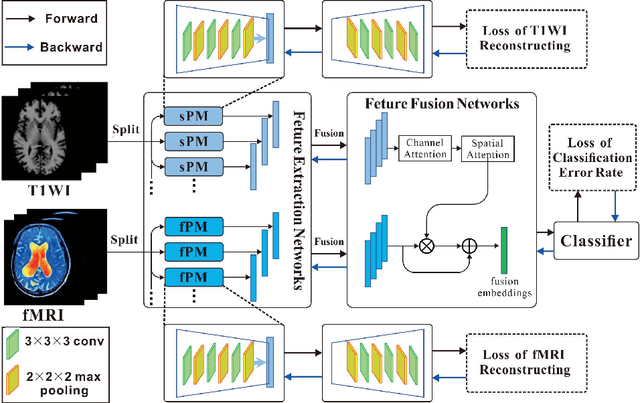

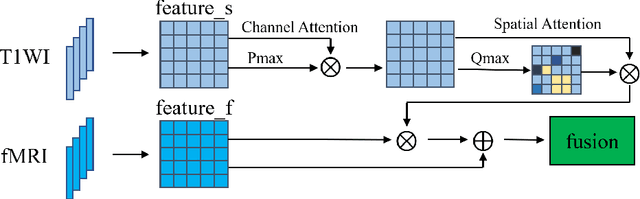
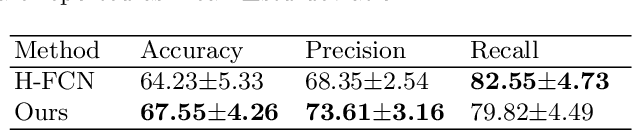
Abstract:The application of machine learning algorithms to the diagnosis and analysis of Alzheimer's disease (AD) from multimodal neuroimaging data is a current research hotspot. It remains a formidable challenge to learn brain region information and discover disease mechanisms from various magnetic resonance images (MRI). In this paper, we propose a simple but highly efficient end-to-end model, a multiscale autoencoder with structural-functional attention network (MASAN) to extract disease-related representations using T1-weighted Imaging (T1WI) and functional MRI (fMRI). Based on the attention mechanism, our model effectively learns the fused features of brain structure and function and finally is trained for the classification of Alzheimer's disease. Compared with the fully convolutional network, the proposed method has further improvement in both accuracy and precision, leading by 3% to 5%. By visualizing the extracted embedding, the empirical results show that there are higher weights on putative AD-related brain regions (such as the hippocampus, amygdala, etc.), and these regions are much more informative in anatomical studies. Conversely, the cerebellum, parietal lobe, thalamus, brain stem, and ventral diencephalon have little predictive contribution.
A Prior Guided Adversarial Representation Learning and Hypergraph Perceptual Network for Predicting Abnormal Connections of Alzheimer's Disease
Oct 12, 2021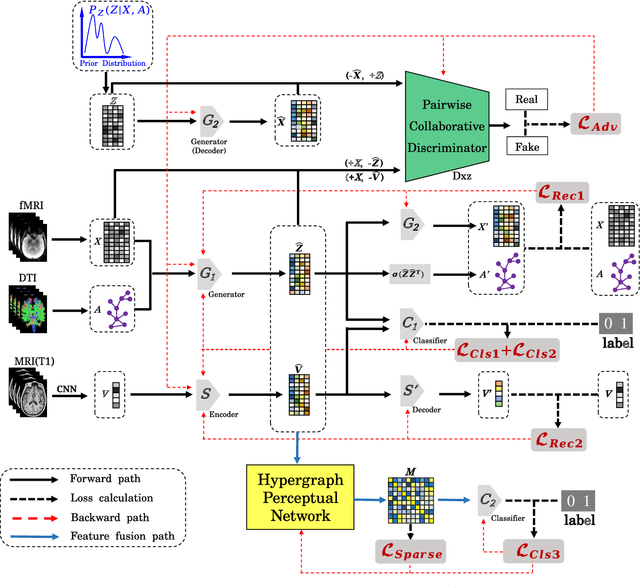
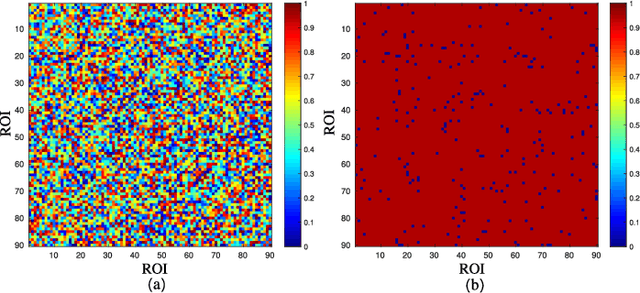
Abstract:Alzheimer's disease is characterized by alterations of the brain's structural and functional connectivity during its progressive degenerative processes. Existing auxiliary diagnostic methods have accomplished the classification task, but few of them can accurately evaluate the changing characteristics of brain connectivity. In this work, a prior guided adversarial representation learning and hypergraph perceptual network (PGARL-HPN) is proposed to predict abnormal brain connections using triple-modality medical images. Concretely, a prior distribution from the anatomical knowledge is estimated to guide multimodal representation learning using an adversarial strategy. Also, the pairwise collaborative discriminator structure is further utilized to narrow the difference of representation distribution. Moreover, the hypergraph perceptual network is developed to effectively fuse the learned representations while establishing high-order relations within and between multimodal images. Experimental results demonstrate that the proposed model outperforms other related methods in analyzing and predicting Alzheimer's disease progression. More importantly, the identified abnormal connections are partly consistent with the previous neuroscience discoveries. The proposed model can evaluate characteristics of abnormal brain connections at different stages of Alzheimer's disease, which is helpful for cognitive disease study and early treatment.
Multimodal Representations Learning and Adversarial Hypergraph Fusion for Early Alzheimer's Disease Prediction
Jul 21, 2021



Abstract:Multimodal neuroimage can provide complementary information about the dementia, but small size of complete multimodal data limits the ability in representation learning. Moreover, the data distribution inconsistency from different modalities may lead to ineffective fusion, which fails to sufficiently explore the intra-modal and inter-modal interactions and compromises the disease diagnosis performance. To solve these problems, we proposed a novel multimodal representation learning and adversarial hypergraph fusion (MRL-AHF) framework for Alzheimer's disease diagnosis using complete trimodal images. First, adversarial strategy and pre-trained model are incorporated into the MRL to extract latent representations from multimodal data. Then two hypergraphs are constructed from the latent representations and the adversarial network based on graph convolution is employed to narrow the distribution difference of hyperedge features. Finally, the hyperedge-invariant features are fused for disease prediction by hyperedge convolution. Experiments on the public Alzheimer's Disease Neuroimaging Initiative(ADNI) database demonstrate that our model achieves superior performance on Alzheimer's disease detection compared with other related models and provides a possible way to understand the underlying mechanisms of disorder's progression by analyzing the abnormal brain connections.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge