Changwei Gong
Multi-Head Spectral-Adaptive Graph Anomaly Detection
Dec 25, 2025Abstract:Graph anomaly detection technology has broad applications in financial fraud and risk control. However, existing graph anomaly detection methods often face significant challenges when dealing with complex and variable abnormal patterns, as anomalous nodes are often disguised and mixed with normal nodes, leading to the coexistence of homophily and heterophily in the graph domain. Recent spectral graph neural networks have made notable progress in addressing this issue; however, current techniques typically employ fixed, globally shared filters. This 'one-size-fits-all' approach can easily cause over-smoothing, erasing critical high-frequency signals needed for fraud detection, and lacks adaptive capabilities for different graph instances. To solve this problem, we propose a Multi-Head Spectral-Adaptive Graph Neural Network (MHSA-GNN). The core innovation is the design of a lightweight hypernetwork that, conditioned on a 'spectral fingerprint' containing structural statistics and Rayleigh quotient features, dynamically generates Chebyshev filter parameters tailored to each instance. This enables a customized filtering strategy for each node and its local subgraph. Additionally, to prevent mode collapse in the multi-head mechanism, we introduce a novel dual regularization strategy that combines teacher-student contrastive learning (TSC) to ensure representation accuracy and Barlow Twins diversity loss (BTD) to enforce orthogonality among heads. Extensive experiments on four real-world datasets demonstrate that our method effectively preserves high-frequency abnormal signals and significantly outperforms existing state-of-the-art methods, especially showing excellent robustness on highly heterogeneous datasets.
UIE-UnFold: Deep Unfolding Network with Color Priors and Vision Transformer for Underwater Image Enhancement
Aug 20, 2024



Abstract:Underwater image enhancement (UIE) plays a crucial role in various marine applications, but it remains challenging due to the complex underwater environment. Current learning-based approaches frequently lack explicit incorporation of prior knowledge about the physical processes involved in underwater image formation, resulting in limited optimization despite their impressive enhancement results. This paper proposes a novel deep unfolding network (DUN) for UIE that integrates color priors and inter-stage feature transformation to improve enhancement performance. The proposed DUN model combines the iterative optimization and reliability of model-based methods with the flexibility and representational power of deep learning, offering a more explainable and stable solution compared to existing learning-based UIE approaches. The proposed model consists of three key components: a Color Prior Guidance Block (CPGB) that establishes a mapping between color channels of degraded and original images, a Nonlinear Activation Gradient Descent Module (NAGDM) that simulates the underwater image degradation process, and an Inter Stage Feature Transformer (ISF-Former) that facilitates feature exchange between different network stages. By explicitly incorporating color priors and modeling the physical characteristics of underwater image formation, the proposed DUN model achieves more accurate and reliable enhancement results. Extensive experiments on multiple underwater image datasets demonstrate the superiority of the proposed model over state-of-the-art methods in both quantitative and qualitative evaluations. The proposed DUN-based approach offers a promising solution for UIE, enabling more accurate and reliable scientific analysis in marine research. The code is available at https://github.com/CXH-Research/UIE-UnFold.
Generative artificial intelligence-enabled dynamic detection of nicotine-related circuits
Dec 13, 2022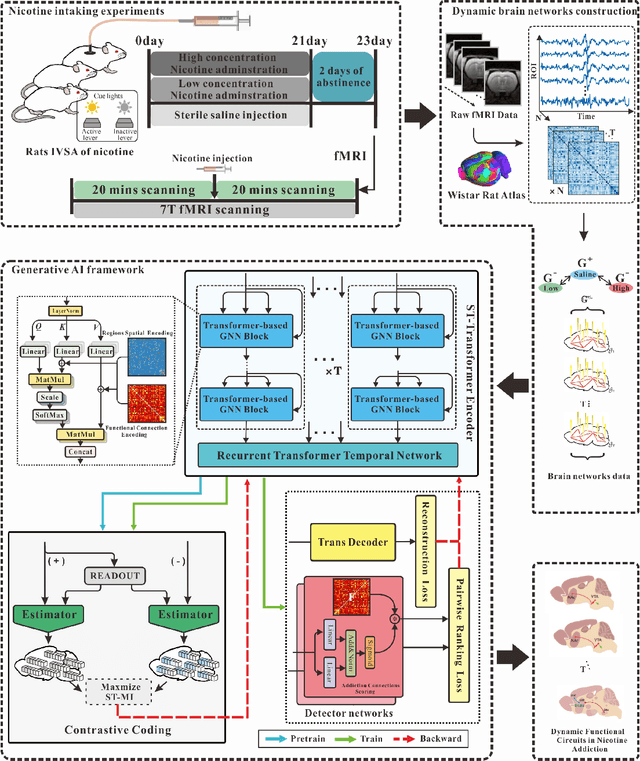
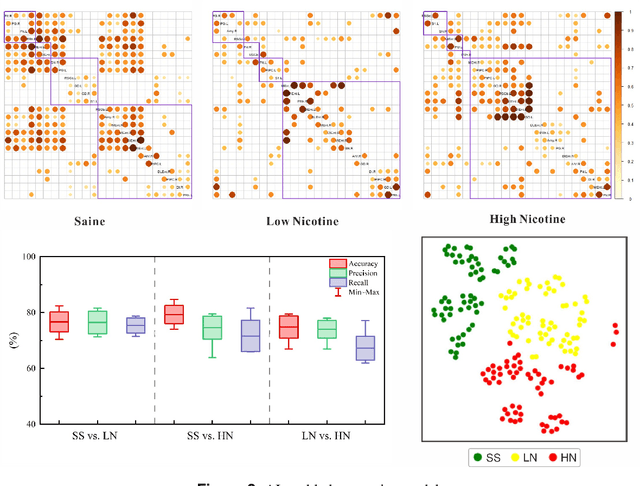

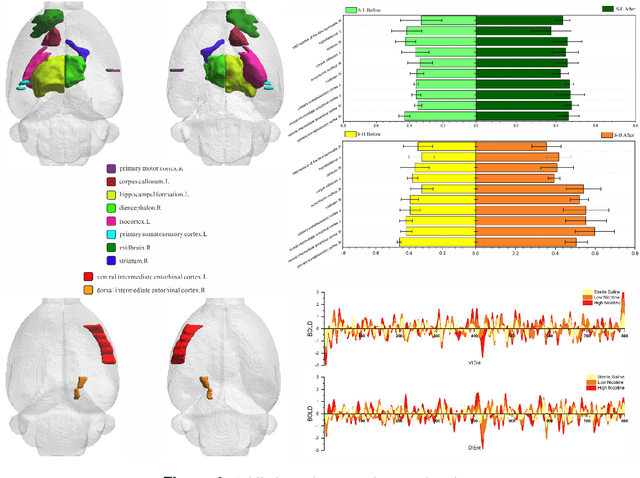
Abstract:The identification of addiction-related circuits is critical for explaining addiction processes and developing addiction treatments. And models of functional addiction circuits developed from functional imaging are an effective tool for discovering and verifying addiction circuits. However, analyzing functional imaging data of addiction and detecting functional addiction circuits still have challenges. We have developed a data-driven and end-to-end generative artificial intelligence(AI) framework to address these difficulties. The framework integrates dynamic brain network modeling and novel network architecture networks architecture, including temporal graph Transformer and contrastive learning modules. A complete workflow is formed by our generative AI framework: the functional imaging data, from neurobiological experiments, and computational modeling, to end-to-end neural networks, is transformed into dynamic nicotine addiction-related circuits. It enables the detection of addiction-related brain circuits with dynamic properties and reveals the underlying mechanisms of addiction.
Feature-selected Graph Spatial Attention Network for Addictive Brain-Networks Identification
Jul 05, 2022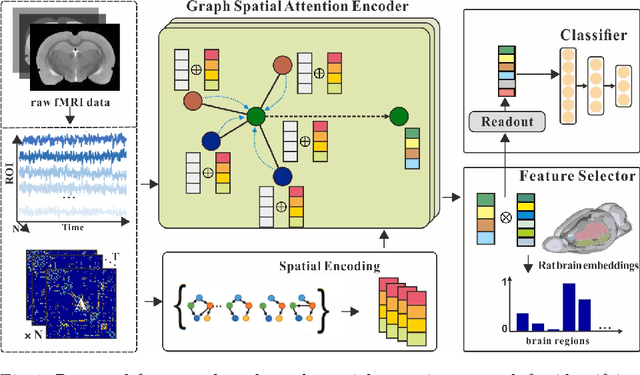

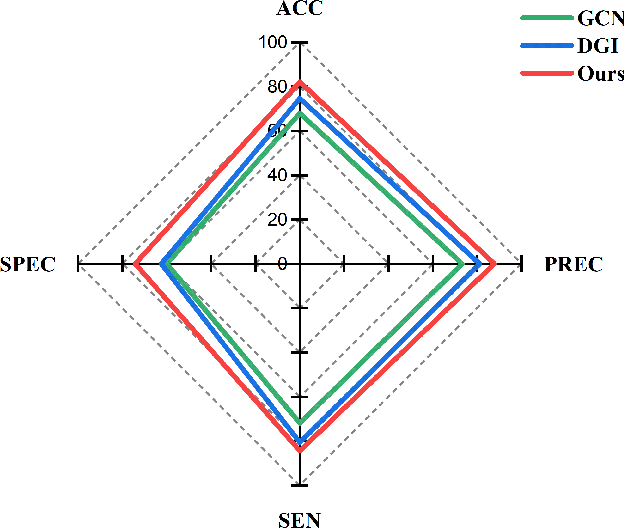
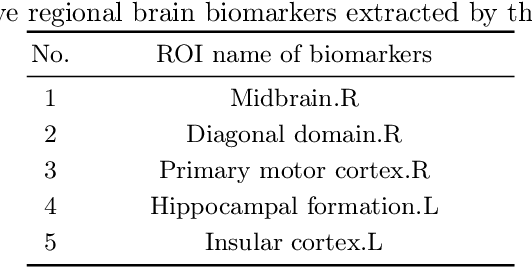
Abstract:Functional alterations in the relevant neural circuits occur from drug addiction over a certain period. And these significant alterations are also revealed by analyzing fMRI. However, because of fMRI's high dimensionality and poor signal-to-noise ratio, it is challenging to encode efficient and robust brain regional embeddings for both graph-level identification and region-level biomarkers detection tasks between nicotine addiction (NA) and healthy control (HC) groups. In this work, we represent the fMRI of the rat brain as a graph with biological attributes and propose a novel feature-selected graph spatial attention network(FGSAN) to extract the biomarkers of addiction and identify from these brain networks. Specially, a graph spatial attention encoder is employed to capture the features of spatiotemporal brain networks with spatial information. The method simultaneously adopts a Bayesian feature selection strategy to optimize the model and improve classification task by constraining features. Experiments on an addiction-related neural imaging dataset show that the proposed model can obtain superior performance and detect interpretable biomarkers associated with addiction-relevant neural circuits.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge