Hao Du
ERNIE 5.0 Technical Report
Feb 04, 2026Abstract:In this report, we introduce ERNIE 5.0, a natively autoregressive foundation model desinged for unified multimodal understanding and generation across text, image, video, and audio. All modalities are trained from scratch under a unified next-group-of-tokens prediction objective, based on an ultra-sparse mixture-of-experts (MoE) architecture with modality-agnostic expert routing. To address practical challenges in large-scale deployment under diverse resource constraints, ERNIE 5.0 adopts a novel elastic training paradigm. Within a single pre-training run, the model learns a family of sub-models with varying depths, expert capacities, and routing sparsity, enabling flexible trade-offs among performance, model size, and inference latency in memory- or time-constrained scenarios. Moreover, we systematically address the challenges of scaling reinforcement learning to unified foundation models, thereby guaranteeing efficient and stable post-training under ultra-sparse MoE architectures and diverse multimodal settings. Extensive experiments demonstrate that ERNIE 5.0 achieves strong and balanced performance across multiple modalities. To the best of our knowledge, among publicly disclosed models, ERNIE 5.0 represents the first production-scale realization of a trillion-parameter unified autoregressive model that supports both multimodal understanding and generation. To facilitate further research, we present detailed visualizations of modality-agnostic expert routing in the unified model, alongside comprehensive empirical analysis of elastic training, aiming to offer profound insights to the community.
Pushing the Limits of Safety: A Technical Report on the ATLAS Challenge 2025
Jun 14, 2025Abstract:Multimodal Large Language Models (MLLMs) have enabled transformative advancements across diverse applications but remain susceptible to safety threats, especially jailbreak attacks that induce harmful outputs. To systematically evaluate and improve their safety, we organized the Adversarial Testing & Large-model Alignment Safety Grand Challenge (ATLAS) 2025}. This technical report presents findings from the competition, which involved 86 teams testing MLLM vulnerabilities via adversarial image-text attacks in two phases: white-box and black-box evaluations. The competition results highlight ongoing challenges in securing MLLMs and provide valuable guidance for developing stronger defense mechanisms. The challenge establishes new benchmarks for MLLM safety evaluation and lays groundwork for advancing safer multimodal AI systems. The code and data for this challenge are openly available at https://github.com/NY1024/ATLAS_Challenge_2025.
Can Differentially Private Fine-tuning LLMs Protect Against Privacy Attacks?
May 01, 2025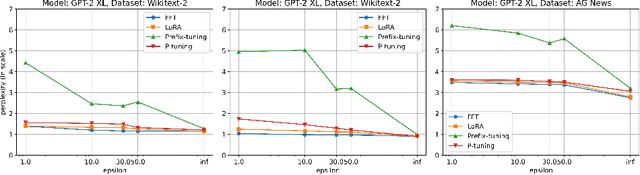
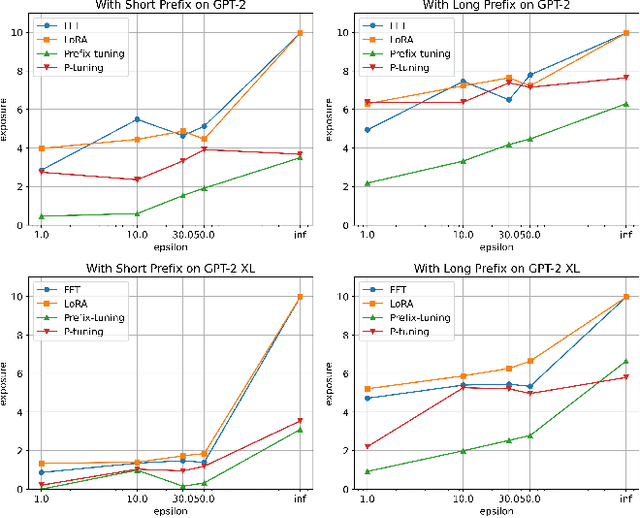
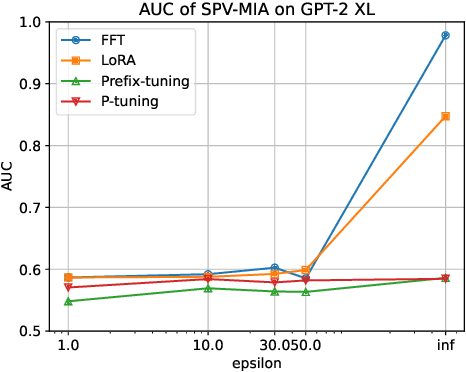
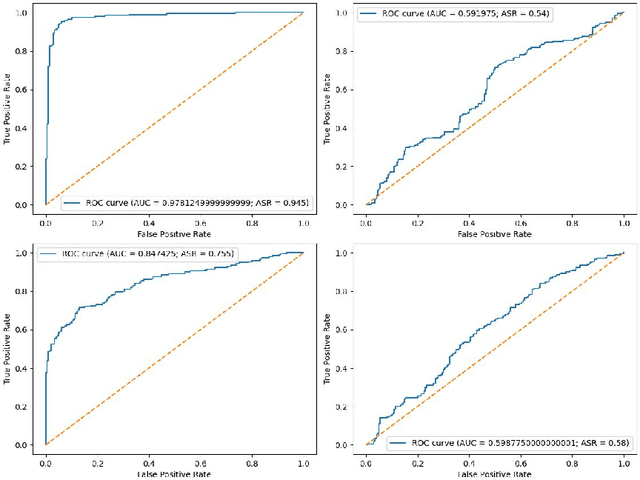
Abstract:Fine-tuning large language models (LLMs) has become an essential strategy for adapting them to specialized tasks; however, this process introduces significant privacy challenges, as sensitive training data may be inadvertently memorized and exposed. Although differential privacy (DP) offers strong theoretical guarantees against such leakage, its empirical privacy effectiveness on LLMs remains unclear, especially under different fine-tuning methods. In this paper, we systematically investigate the impact of DP across fine-tuning methods and privacy budgets, using both data extraction and membership inference attacks to assess empirical privacy risks. Our main findings are as follows: (1) Differential privacy reduces model utility, but its impact varies significantly across different fine-tuning methods. (2) Without DP, the privacy risks of models fine-tuned with different approaches differ considerably. (3) When DP is applied, even a relatively high privacy budget can substantially lower privacy risk. (4) The privacy-utility trade-off under DP training differs greatly among fine-tuning methods, with some methods being unsuitable for DP due to severe utility degradation. Our results provide practical guidance for privacy-conscious deployment of LLMs and pave the way for future research on optimizing the privacy-utility trade-off in fine-tuning methodologies.
SVLTA: Benchmarking Vision-Language Temporal Alignment via Synthetic Video Situation
Apr 08, 2025Abstract:Vision-language temporal alignment is a crucial capability for human dynamic recognition and cognition in real-world scenarios. While existing research focuses on capturing vision-language relevance, it faces limitations due to biased temporal distributions, imprecise annotations, and insufficient compositionally. To achieve fair evaluation and comprehensive exploration, our objective is to investigate and evaluate the ability of models to achieve alignment from a temporal perspective, specifically focusing on their capacity to synchronize visual scenarios with linguistic context in a temporally coherent manner. As a preliminary step, we present the statistical analysis of existing benchmarks and reveal the existing challenges from a decomposed perspective. To this end, we introduce SVLTA, the Synthetic Vision-Language Temporal Alignment derived via a well-designed and feasible control generation method within a simulation environment. The approach considers commonsense knowledge, manipulable action, and constrained filtering, which generates reasonable, diverse, and balanced data distributions for diagnostic evaluations. Our experiments reveal diagnostic insights through the evaluations in temporal question answering, distributional shift sensitiveness, and temporal alignment adaptation.
Tumor Detection, Segmentation and Classification Challenge on Automated 3D Breast Ultrasound: The TDSC-ABUS Challenge
Jan 26, 2025



Abstract:Breast cancer is one of the most common causes of death among women worldwide. Early detection helps in reducing the number of deaths. Automated 3D Breast Ultrasound (ABUS) is a newer approach for breast screening, which has many advantages over handheld mammography such as safety, speed, and higher detection rate of breast cancer. Tumor detection, segmentation, and classification are key components in the analysis of medical images, especially challenging in the context of 3D ABUS due to the significant variability in tumor size and shape, unclear tumor boundaries, and a low signal-to-noise ratio. The lack of publicly accessible, well-labeled ABUS datasets further hinders the advancement of systems for breast tumor analysis. Addressing this gap, we have organized the inaugural Tumor Detection, Segmentation, and Classification Challenge on Automated 3D Breast Ultrasound 2023 (TDSC-ABUS2023). This initiative aims to spearhead research in this field and create a definitive benchmark for tasks associated with 3D ABUS image analysis. In this paper, we summarize the top-performing algorithms from the challenge and provide critical analysis for ABUS image examination. We offer the TDSC-ABUS challenge as an open-access platform at https://tdsc-abus2023.grand-challenge.org/ to benchmark and inspire future developments in algorithmic research.
Privacy in Fine-tuning Large Language Models: Attacks, Defenses, and Future Directions
Dec 21, 2024


Abstract:Fine-tuning has emerged as a critical process in leveraging Large Language Models (LLMs) for specific downstream tasks, enabling these models to achieve state-of-the-art performance across various domains. However, the fine-tuning process often involves sensitive datasets, introducing privacy risks that exploit the unique characteristics of this stage. In this paper, we provide a comprehensive survey of privacy challenges associated with fine-tuning LLMs, highlighting vulnerabilities to various privacy attacks, including membership inference, data extraction, and backdoor attacks. We further review defense mechanisms designed to mitigate privacy risks in the fine-tuning phase, such as differential privacy, federated learning, and knowledge unlearning, discussing their effectiveness and limitations in addressing privacy risks and maintaining model utility. By identifying key gaps in existing research, we highlight challenges and propose directions to advance the development of privacy-preserving methods for fine-tuning LLMs, promoting their responsible use in diverse applications.
Preserving Tumor Volumes for Unsupervised Medical Image Registration
Sep 18, 2023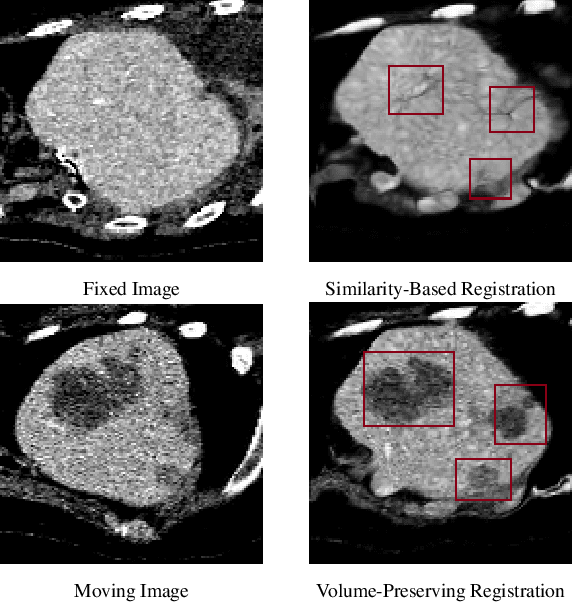
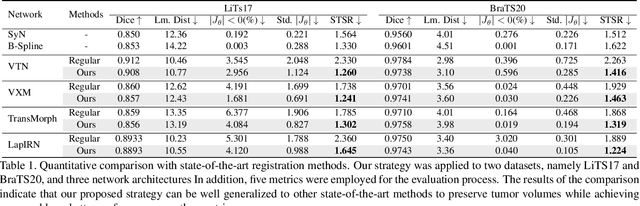
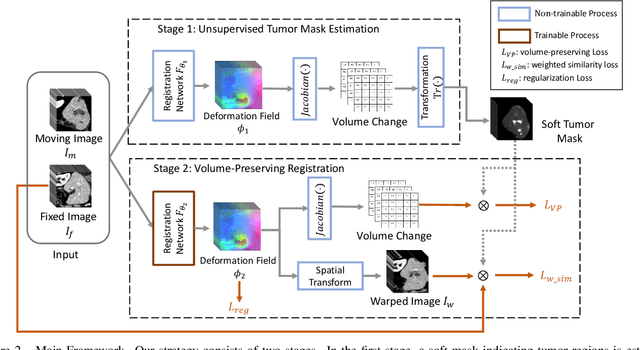
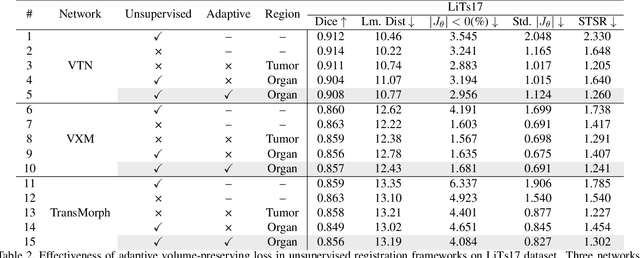
Abstract:Medical image registration is a critical task that estimates the spatial correspondence between pairs of images. However, current traditional and deep-learning-based methods rely on similarity measures to generate a deforming field, which often results in disproportionate volume changes in dissimilar regions, especially in tumor regions. These changes can significantly alter the tumor size and underlying anatomy, which limits the practical use of image registration in clinical diagnosis. To address this issue, we have formulated image registration with tumors as a constraint problem that preserves tumor volumes while maximizing image similarity in other normal regions. Our proposed strategy involves a two-stage process. In the first stage, we use similarity-based registration to identify potential tumor regions by their volume change, generating a soft tumor mask accordingly. In the second stage, we propose a volume-preserving registration with a novel adaptive volume-preserving loss that penalizes the change in size adaptively based on the masks calculated from the previous stage. Our approach balances image similarity and volume preservation in different regions, i.e., normal and tumor regions, by using soft tumor masks to adjust the imposition of volume-preserving loss on each one. This ensures that the tumor volume is preserved during the registration process. We have evaluated our strategy on various datasets and network architectures, demonstrating that our method successfully preserves the tumor volume while achieving comparable registration results with state-of-the-art methods. Our codes is available at: \url{https://dddraxxx.github.io/Volume-Preserving-Registration/}.
Weakly-Supervised 3D Medical Image Segmentation using Geometric Prior and Contrastive Similarity
Feb 04, 2023Abstract:Medical image segmentation is almost the most important pre-processing procedure in computer-aided diagnosis but is also a very challenging task due to the complex shapes of segments and various artifacts caused by medical imaging, (i.e., low-contrast tissues, and non-homogenous textures). In this paper, we propose a simple yet effective segmentation framework that incorporates the geometric prior and contrastive similarity into the weakly-supervised segmentation framework in a loss-based fashion. The proposed geometric prior built on point cloud provides meticulous geometry to the weakly-supervised segmentation proposal, which serves as better supervision than the inherent property of the bounding-box annotation (i.e., height and width). Furthermore, we propose contrastive similarity to encourage organ pixels to gather around in the contrastive embedding space, which helps better distinguish low-contrast tissues. The proposed contrastive embedding space can make up for the poor representation of the conventionally-used gray space. Extensive experiments are conducted to verify the effectiveness and the robustness of the proposed weakly-supervised segmentation framework. The proposed framework is superior to state-of-the-art weakly-supervised methods on the following publicly accessible datasets: LiTS 2017 Challenge, KiTS 2021 Challenge, and LPBA40. We also dissect our method and evaluate the performance of each component.
Deep Portrait Lighting Enhancement with 3D Guidance
Aug 04, 2021Abstract:Despite recent breakthroughs in deep learning methods for image lighting enhancement, they are inferior when applied to portraits because 3D facial information is ignored in their models. To address this, we present a novel deep learning framework for portrait lighting enhancement based on 3D facial guidance. Our framework consists of two stages. In the first stage, corrected lighting parameters are predicted by a network from the input bad lighting image, with the assistance of a 3D morphable model and a differentiable renderer. Given the predicted lighting parameter, the differentiable renderer renders a face image with corrected shading and texture, which serves as the 3D guidance for learning image lighting enhancement in the second stage. To better exploit the long-range correlations between the input and the guidance, in the second stage, we design an image-to-image translation network with a novel transformer architecture, which automatically produces a lighting-enhanced result. Experimental results on the FFHQ dataset and in-the-wild images show that the proposed method outperforms state-of-the-art methods in terms of both quantitative metrics and visual quality. We will publish our dataset along with more results on https://cassiepython.github.io/egsr/index.html.
* {\dag} for equal conribution. Accepted to CGF. Project page: https://cassiepython.github.io/egsr/index.html
Multi-task Graph Convolutional Neural Network for Calcification Morphology and Distribution Analysis in Mammograms
May 14, 2021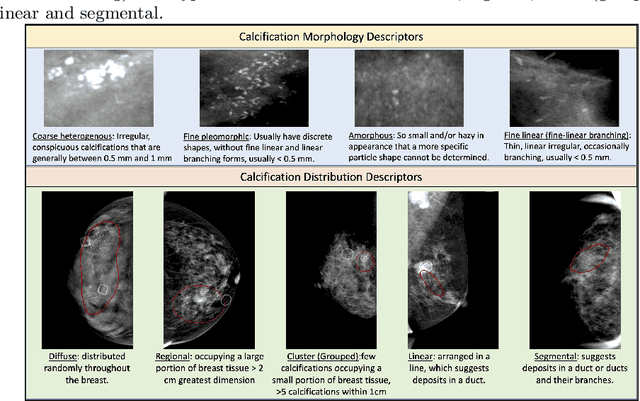
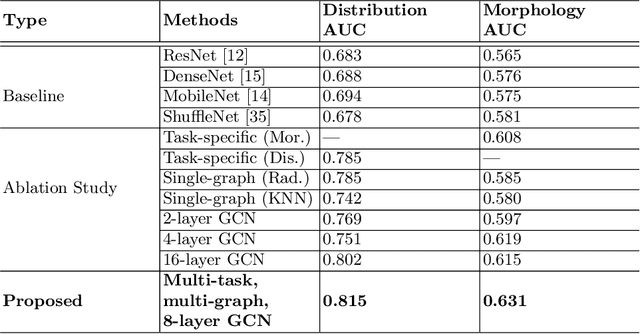
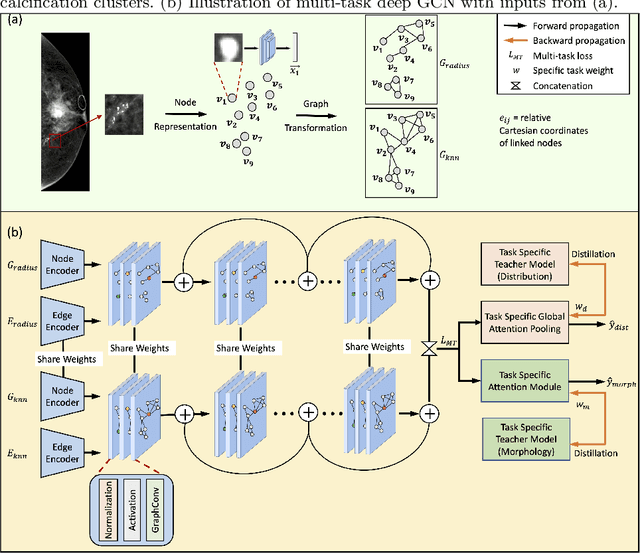
Abstract:The morphology and distribution of microcalcifications in a cluster are the most important characteristics for radiologists to diagnose breast cancer. However, it is time-consuming and difficult for radiologists to identify these characteristics, and there also lacks of effective solutions for automatic characterization. In this study, we proposed a multi-task deep graph convolutional network (GCN) method for the automatic characterization of morphology and distribution of microcalcifications in mammograms. Our proposed method transforms morphology and distribution characterization into node and graph classification problem and learns the representations concurrently. Through extensive experiments, we demonstrate significant improvements with the proposed multi-task GCN comparing to the baselines. Moreover, the achieved improvements can be related to and enhance clinical understandings. We explore, for the first time, the application of GCNs in microcalcification characterization that suggests the potential of graph learning for more robust understanding of medical images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge