Ying Song
GraphToxin: Reconstructing Full Unlearned Graphs from Graph Unlearning
Nov 14, 2025



Abstract:Graph unlearning has emerged as a promising solution for complying with "the right to be forgotten" regulations by enabling the removal of sensitive information upon request. However, this solution is not foolproof. The involvement of multiple parties creates new attack surfaces, and residual traces of deleted data can still remain in the unlearned graph neural networks. These vulnerabilities can be exploited by attackers to recover the supposedly erased samples, thereby undermining the inherent functionality of graph unlearning. In this work, we propose GraphToxin, the first graph reconstruction attack against graph unlearning. Specifically, we introduce a novel curvature matching module to provide a fine-grained guidance for full unlearned graph recovery. We demonstrate that GraphToxin can successfully subvert the regulatory guarantees expected from graph unlearning - it can recover not only a deleted individual's information and personal links but also sensitive content from their connections, thereby posing substantially more detrimental threats. Furthermore, we extend GraphToxin to multiple node removals under both white-box and black-box setting. We highlight the necessity of a worst-case analysis and propose a comprehensive evaluation framework to systematically assess the attack performance under both random and worst-case node removals. This provides a more robust and realistic measure of the vulnerability of graph unlearning methods to graph reconstruction attacks. Our extensive experiments demonstrate the effectiveness and flexibility of GraphToxin. Notably, we show that existing defense mechanisms are largely ineffective against this attack and, in some cases, can even amplify its performance. Given the severe privacy risks posed by GraphToxin, our work underscores the urgent need for the development of more effective and robust defense strategies against this attack.
Class Incremental Medical Image Segmentation via Prototype-Guided Calibration and Dual-Aligned Distillation
Nov 11, 2025



Abstract:Class incremental medical image segmentation (CIMIS) aims to preserve knowledge of previously learned classes while learning new ones without relying on old-class labels. However, existing methods 1) either adopt one-size-fits-all strategies that treat all spatial regions and feature channels equally, which may hinder the preservation of accurate old knowledge, 2) or focus solely on aligning local prototypes with global ones for old classes while overlooking their local representations in new data, leading to knowledge degradation. To mitigate the above issues, we propose Prototype-Guided Calibration Distillation (PGCD) and Dual-Aligned Prototype Distillation (DAPD) for CIMIS in this paper. Specifically, PGCD exploits prototype-to-feature similarity to calibrate class-specific distillation intensity in different spatial regions, effectively reinforcing reliable old knowledge and suppressing misleading information from old classes. Complementarily, DAPD aligns the local prototypes of old classes extracted from the current model with both global prototypes and local prototypes, further enhancing segmentation performance on old categories. Comprehensive evaluations on two widely used multi-organ segmentation benchmarks demonstrate that our method outperforms state-of-the-art methods, highlighting its robustness and generalization capabilities.
Exploiting Unlabeled Structures through Task Consistency Training for Versatile Medical Image Segmentation
Sep 05, 2025



Abstract:Versatile medical image segmentation (VMIS) targets the segmentation of multiple classes, while obtaining full annotations for all classes is often impractical due to the time and labor required. Leveraging partially labeled datasets (PLDs) presents a promising alternative; however, current VMIS approaches face significant class imbalance due to the unequal category distribution in PLDs. Existing methods attempt to address this by generating pseudo-full labels. Nevertheless, these typically require additional models and often result in potential performance degradation from label noise. In this work, we introduce a Task Consistency Training (TCT) framework to address class imbalance without requiring extra models. TCT includes a backbone network with a main segmentation head (MSH) for multi-channel predictions and multiple auxiliary task heads (ATHs) for task-specific predictions. By enforcing a consistency constraint between the MSH and ATH predictions, TCT effectively utilizes unlabeled anatomical structures. To avoid error propagation from low-consistency, potentially noisy data, we propose a filtering strategy to exclude such data. Additionally, we introduce a unified auxiliary uncertainty-weighted loss (UAUWL) to mitigate segmentation quality declines caused by the dominance of specific tasks. Extensive experiments on eight abdominal datasets from diverse clinical sites demonstrate our approach's effectiveness.
Krait: A Backdoor Attack Against Graph Prompt Tuning
Jul 18, 2024Abstract:Graph prompt tuning has emerged as a promising paradigm to effectively transfer general graph knowledge from pre-trained models to various downstream tasks, particularly in few-shot contexts. However, its susceptibility to backdoor attacks, where adversaries insert triggers to manipulate outcomes, raises a critical concern. We conduct the first study to investigate such vulnerability, revealing that backdoors can disguise benign graph prompts, thus evading detection. We introduce Krait, a novel graph prompt backdoor. Specifically, we propose a simple yet effective model-agnostic metric called label non-uniformity homophily to select poisoned candidates, significantly reducing computational complexity. To accommodate diverse attack scenarios and advanced attack types, we design three customizable trigger generation methods to craft prompts as triggers. We propose a novel centroid similarity-based loss function to optimize prompt tuning for attack effectiveness and stealthiness. Experiments on four real-world graphs demonstrate that Krait can efficiently embed triggers to merely 0.15% to 2% of training nodes, achieving high attack success rates without sacrificing clean accuracy. Notably, in one-to-one and all-to-one attacks, Krait can achieve 100% attack success rates by poisoning as few as 2 and 22 nodes, respectively. Our experiments further show that Krait remains potent across different transfer cases, attack types, and graph neural network backbones. Additionally, Krait can be successfully extended to the black-box setting, posing more severe threats. Finally, we analyze why Krait can evade both classical and state-of-the-art defenses, and provide practical insights for detecting and mitigating this class of attacks.
MAPPING: Debiasing Graph Neural Networks for Fair Node Classification with Limited Sensitive Information Leakage
Jan 23, 2024Abstract:Despite remarkable success in diverse web-based applications, Graph Neural Networks(GNNs) inherit and further exacerbate historical discrimination and social stereotypes, which critically hinder their deployments in high-stake domains such as online clinical diagnosis, financial crediting, etc. However, current fairness research that primarily craft on i.i.d data, cannot be trivially replicated to non-i.i.d. graph structures with topological dependence among samples. Existing fair graph learning typically favors pairwise constraints to achieve fairness but fails to cast off dimensional limitations and generalize them into multiple sensitive attributes; besides, most studies focus on in-processing techniques to enforce and calibrate fairness, constructing a model-agnostic debiasing GNN framework at the pre-processing stage to prevent downstream misuses and improve training reliability is still largely under-explored. Furthermore, previous work on GNNs tend to enhance either fairness or privacy individually but few probe into their interplays. In this paper, we propose a novel model-agnostic debiasing framework named MAPPING (\underline{M}asking \underline{A}nd \underline{P}runing and Message-\underline{P}assing train\underline{ING}) for fair node classification, in which we adopt the distance covariance($dCov$)-based fairness constraints to simultaneously reduce feature and topology biases in arbitrary dimensions, and combine them with adversarial debiasing to confine the risks of attribute inference attacks. Experiments on real-world datasets with different GNN variants demonstrate the effectiveness and flexibility of MAPPING. Our results show that MAPPING can achieve better trade-offs between utility and fairness, and mitigate privacy risks of sensitive information leakage.
Preserving Tumor Volumes for Unsupervised Medical Image Registration
Sep 18, 2023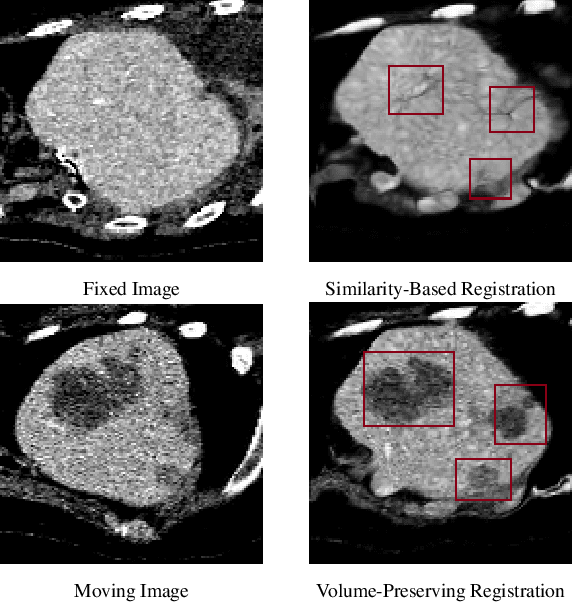
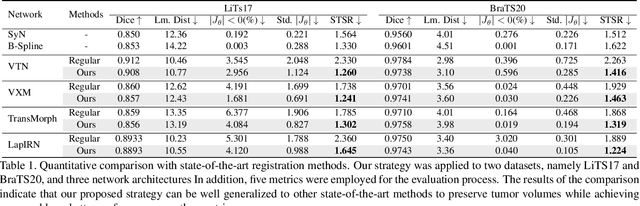
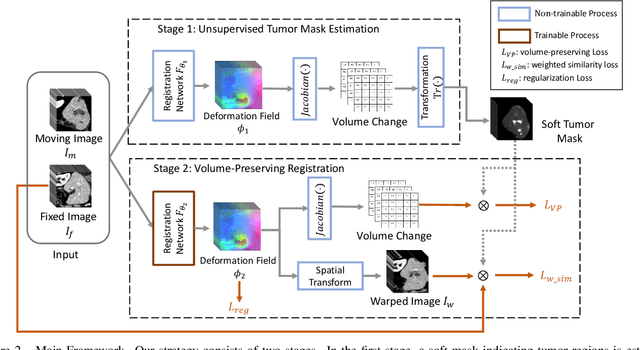
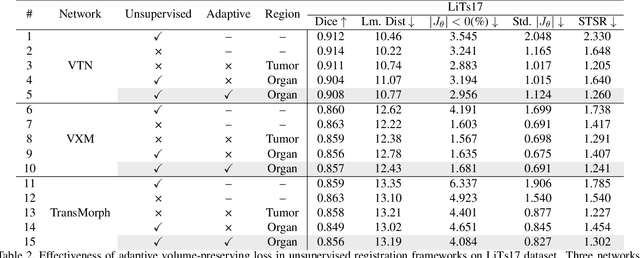
Abstract:Medical image registration is a critical task that estimates the spatial correspondence between pairs of images. However, current traditional and deep-learning-based methods rely on similarity measures to generate a deforming field, which often results in disproportionate volume changes in dissimilar regions, especially in tumor regions. These changes can significantly alter the tumor size and underlying anatomy, which limits the practical use of image registration in clinical diagnosis. To address this issue, we have formulated image registration with tumors as a constraint problem that preserves tumor volumes while maximizing image similarity in other normal regions. Our proposed strategy involves a two-stage process. In the first stage, we use similarity-based registration to identify potential tumor regions by their volume change, generating a soft tumor mask accordingly. In the second stage, we propose a volume-preserving registration with a novel adaptive volume-preserving loss that penalizes the change in size adaptively based on the masks calculated from the previous stage. Our approach balances image similarity and volume preservation in different regions, i.e., normal and tumor regions, by using soft tumor masks to adjust the imposition of volume-preserving loss on each one. This ensures that the tumor volume is preserved during the registration process. We have evaluated our strategy on various datasets and network architectures, demonstrating that our method successfully preserves the tumor volume while achieving comparable registration results with state-of-the-art methods. Our codes is available at: \url{https://dddraxxx.github.io/Volume-Preserving-Registration/}.
Multi-Grained Angle Representation for Remote Sensing Object Detection
Sep 07, 2022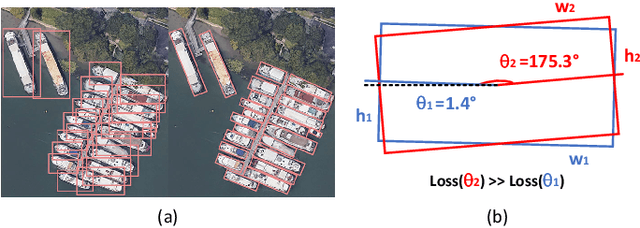
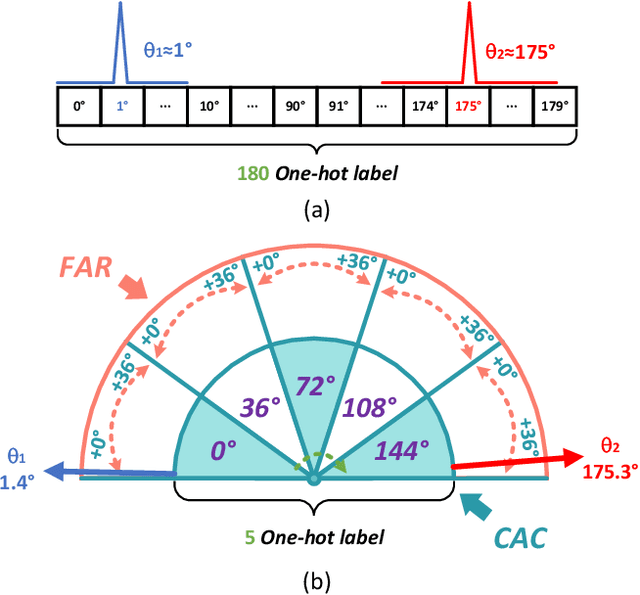
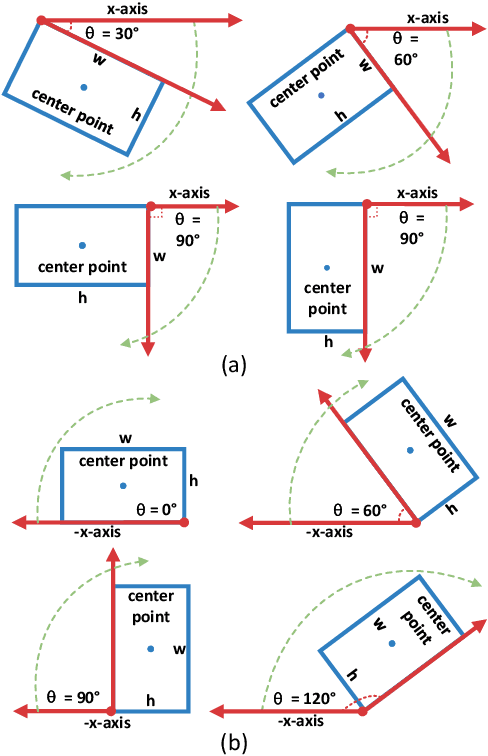
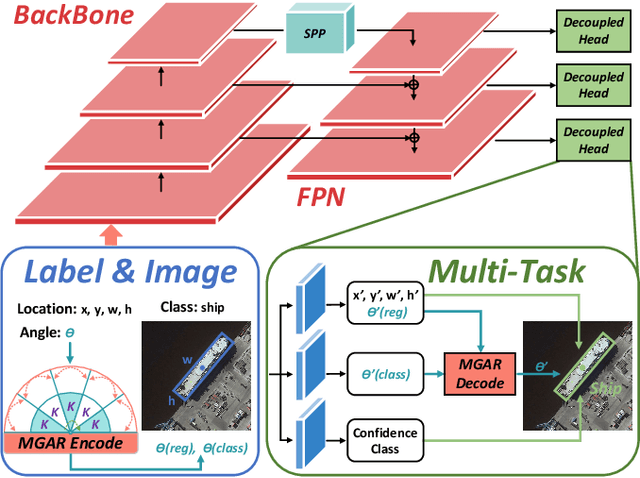
Abstract:Arbitrary-oriented object detection (AOOD) plays a significant role for image understanding in remote sensing scenarios. The existing AOOD methods face the challenges of ambiguity and high costs in angle representation. To this end, a multi-grained angle representation (MGAR) method, consisting of coarse-grained angle classification (CAC) and fine-grained angle regression (FAR), is proposed. Specifically, the designed CAC avoids the ambiguity of angle prediction by discrete angular encoding (DAE) and reduces complexity by coarsening the granularity of DAE. Based on CAC, FAR is developed to refine the angle prediction with much lower costs than narrowing the granularity of DAE. Furthermore, an Intersection over Union (IoU) aware FAR-Loss (IFL) is designed to improve accuracy of angle prediction using an adaptive re-weighting mechanism guided by IoU. Extensive experiments are performed on several public remote sensing datasets, which demonstrate the effectiveness of the proposed MGAR. Moreover, experiments on embedded devices demonstrate that the proposed MGAR is also friendly for lightweight deployments.
Molecular Attributes Transfer from Non-Parallel Data
Nov 30, 2021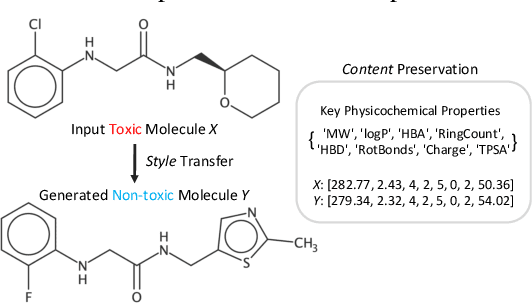
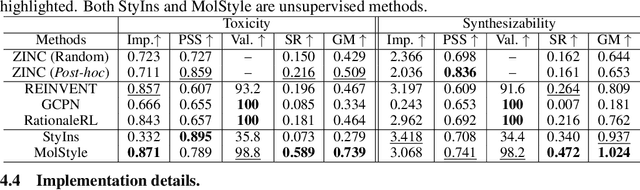
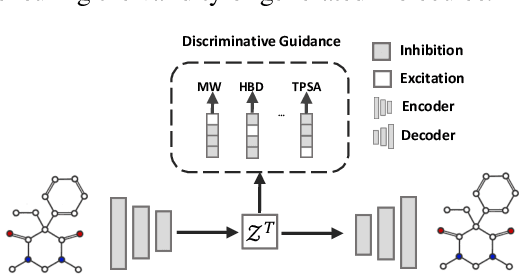
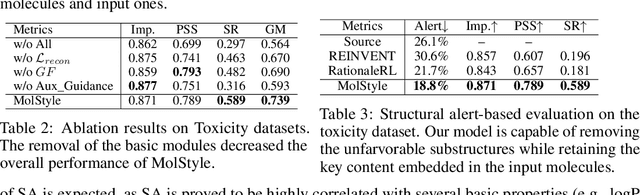
Abstract:Optimizing chemical molecules for desired properties lies at the core of drug development. Despite initial successes made by deep generative models and reinforcement learning methods, these methods were mostly limited by the requirement of predefined attribute functions or parallel data with manually pre-compiled pairs of original and optimized molecules. In this paper, for the first time, we formulate molecular optimization as a style transfer problem and present a novel generative model that could automatically learn internal differences between two groups of non-parallel data through adversarial training strategies. Our model further enables both preservation of molecular contents and optimization of molecular properties through combining auxiliary guided-variational autoencoders and generative flow techniques. Experiments on two molecular optimization tasks, toxicity modification and synthesizability improvement, demonstrate that our model significantly outperforms several state-of-the-art methods.
Learning Attributed Graph Representations with Communicative Message Passing Transformer
Jul 28, 2021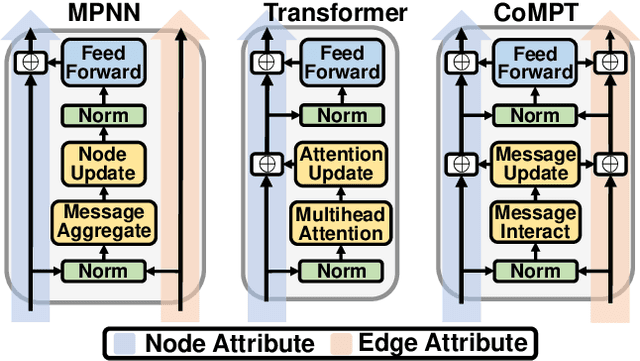
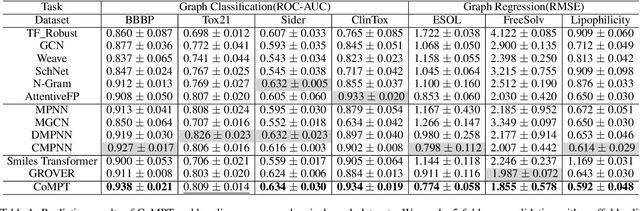
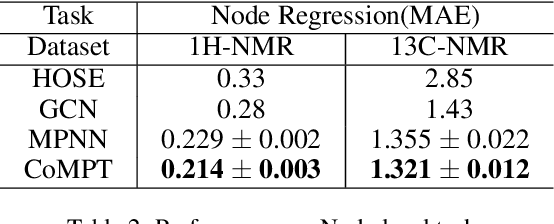
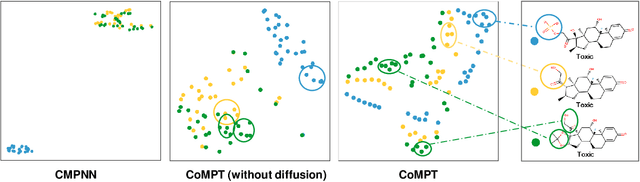
Abstract:Constructing appropriate representations of molecules lies at the core of numerous tasks such as material science, chemistry and drug designs. Recent researches abstract molecules as attributed graphs and employ graph neural networks (GNN) for molecular representation learning, which have made remarkable achievements in molecular graph modeling. Albeit powerful, current models either are based on local aggregation operations and thus miss higher-order graph properties or focus on only node information without fully using the edge information. For this sake, we propose a Communicative Message Passing Transformer (CoMPT) neural network to improve the molecular graph representation by reinforcing message interactions between nodes and edges based on the Transformer architecture. Unlike the previous transformer-style GNNs that treat molecules as fully connected graphs, we introduce a message diffusion mechanism to leverage the graph connectivity inductive bias and reduce the message enrichment explosion. Extensive experiments demonstrated that the proposed model obtained superior performances (around 4$\%$ on average) against state-of-the-art baselines on seven chemical property datasets (graph-level tasks) and two chemical shift datasets (node-level tasks). Further visualization studies also indicated a better representation capacity achieved by our model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge