Jiahua Rao
De Novo Molecular Generation from Mass Spectra via Many-Body Enhanced Diffusion
Feb 02, 2026Abstract:Molecular structure generation from mass spectrometry is fundamental for understanding cellular metabolism and discovering novel compounds. Although tandem mass spectrometry (MS/MS) enables the high-throughput acquisition of fragment fingerprints, these spectra often reflect higher-order interactions involving the concerted cleavage of multiple atoms and bonds-crucial for resolving complex isomers and non-local fragmentation mechanisms. However, most existing methods adopt atom-centric and pairwise interaction modeling, overlooking higher-order edge interactions and lacking the capacity to systematically capture essential many-body characteristics for structure generation. To overcome these limitations, we present MBGen, a Many-Body enhanced diffusion framework for de novo molecular structure Generation from mass spectra. By integrating a many-body attention mechanism and higher-order edge modeling, MBGen comprehensively leverages the rich structural information encoded in MS/MS spectra, enabling accurate de novo generation and isomer differentiation for novel molecules. Experimental results on the NPLIB1 and MassSpecGym benchmarks demonstrate that MBGen achieves superior performance, with improvements of up to 230% over state-of-the-art methods, highlighting the scientific value and practical utility of many-body modeling for mass spectrometry-based molecular generation. Further analysis and ablation studies show that our approach effectively captures higher-order interactions and exhibits enhanced sensitivity to complex isomeric and non-local fragmentation information.
A 3D pocket-aware and affinity-guided diffusion model for lead optimization
Apr 29, 2025Abstract:Molecular optimization, aimed at improving binding affinity or other molecular properties, is a crucial task in drug discovery that often relies on the expertise of medicinal chemists. Recently, deep learning-based 3D generative models showed promise in enhancing the efficiency of molecular optimization. However, these models often struggle to adequately consider binding affinities with protein targets during lead optimization. Herein, we propose a 3D pocket-aware and affinity-guided diffusion model, named Diffleop, to optimize molecules with enhanced binding affinity. The model explicitly incorporates the knowledge of protein-ligand binding affinity to guide the denoising sampling for molecule generation with high affinity. The comprehensive evaluations indicated that Diffleop outperforms baseline models across multiple metrics, especially in terms of binding affinity.
Incorporating Retrieval-based Causal Learning with Information Bottlenecks for Interpretable Graph Neural Networks
Feb 07, 2024



Abstract:Graph Neural Networks (GNNs) have gained considerable traction for their capability to effectively process topological data, yet their interpretability remains a critical concern. Current interpretation methods are dominated by post-hoc explanations to provide a transparent and intuitive understanding of GNNs. However, they have limited performance in interpreting complicated subgraphs and can't utilize the explanation to advance GNN predictions. On the other hand, transparent GNN models are proposed to capture critical subgraphs. While such methods could improve GNN predictions, they usually don't perform well on explanations. Thus, it is desired for a new strategy to better couple GNN explanation and prediction. In this study, we have developed a novel interpretable causal GNN framework that incorporates retrieval-based causal learning with Graph Information Bottleneck (GIB) theory. The framework could semi-parametrically retrieve crucial subgraphs detected by GIB and compress the explanatory subgraphs via a causal module. The framework was demonstrated to consistently outperform state-of-the-art methods, and to achieve 32.71\% higher precision on real-world explanation scenarios with diverse explanation types. More importantly, the learned explanations were shown able to also improve GNN prediction performance.
Retrieval-based Knowledge Augmented Vision Language Pre-training
Apr 27, 2023Abstract:With recent progress in large-scale vision and language representation learning, Vision Language Pretraining (VLP) models have achieved promising improvements on various multi-modal downstream tasks. Albeit powerful, these pre-training models still do not take advantage of world knowledge, which is implicit in multi-modal data but comprises abundant and complementary information. In this work, we propose a REtrieval-based knowledge Augmented Vision Language Pre-training model (REAVL), which retrieves world knowledge from knowledge graphs (KGs) and incorporates them in vision-language pre-training. REAVL has two core components: a knowledge retriever that retrieves knowledge given multi-modal data, and a knowledge-augmented model that fuses multi-modal data and knowledge. By novelly unifying four knowledge-aware self-supervised tasks, REAVL promotes the mutual integration of multi-modal data and knowledge by fusing explicit knowledge with vision-language pairs for masked multi-modal data modeling and KG relational reasoning. Empirical experiments show that REAVL achieves new state-of-the-art performance uniformly on knowledge-based vision-language understanding and multimodal entity linking tasks, and competitive results on general vision-language tasks while only using 0.2% pre-training data of the best models.
Communicative Subgraph Representation Learning for Multi-Relational Inductive Drug-Gene Interaction Prediction
May 12, 2022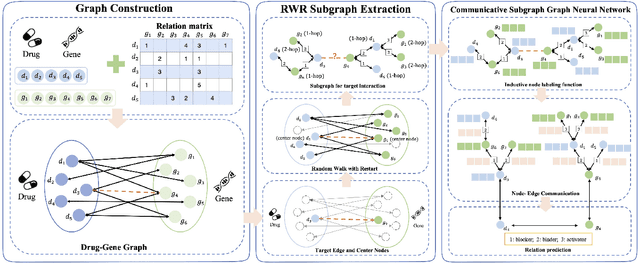
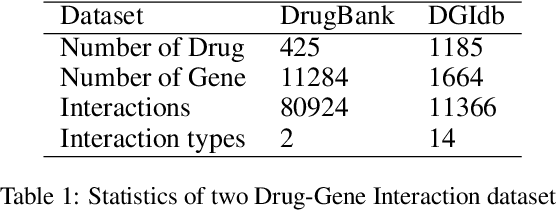
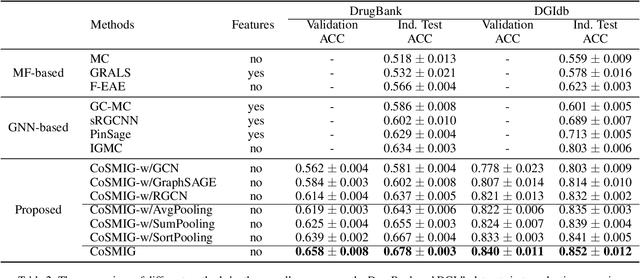
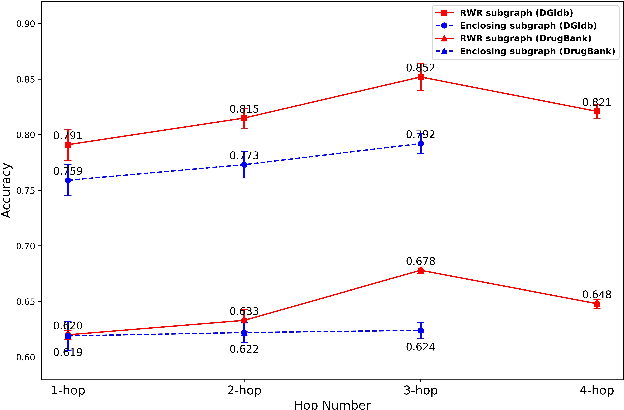
Abstract:Illuminating the interconnections between drugs and genes is an important topic in drug development and precision medicine. Currently, computational predictions of drug-gene interactions mainly focus on the binding interactions without considering other relation types like agonist, antagonist, etc. In addition, existing methods either heavily rely on high-quality domain features or are intrinsically transductive, which limits the capacity of models to generalize to drugs/genes that lack external information or are unseen during the training process. To address these problems, we propose a novel Communicative Subgraph representation learning for Multi-relational Inductive drug-Gene interactions prediction (CoSMIG), where the predictions of drug-gene relations are made through subgraph patterns, and thus are naturally inductive for unseen drugs/genes without retraining or utilizing external domain features. Moreover, the model strengthened the relations on the drug-gene graph through a communicative message passing mechanism. To evaluate our method, we compiled two new benchmark datasets from DrugBank and DGIdb. The comprehensive experiments on the two datasets showed that our method outperformed state-of-the-art baselines in the transductive scenarios and achieved superior performance in the inductive ones. Further experimental analysis including LINCS experimental validation and literature verification also demonstrated the value of our model.
Learning Attributed Graph Representations with Communicative Message Passing Transformer
Jul 28, 2021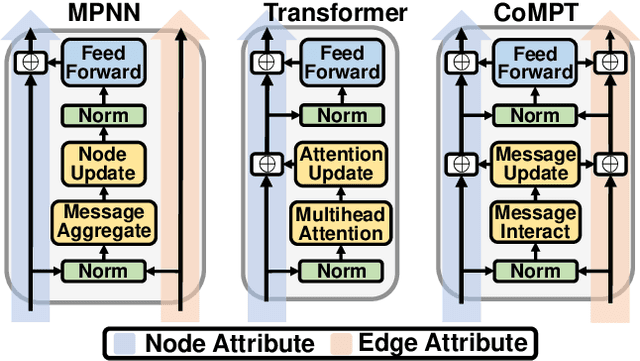
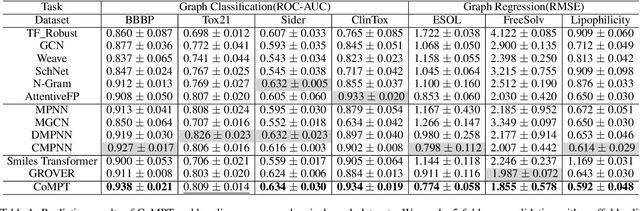
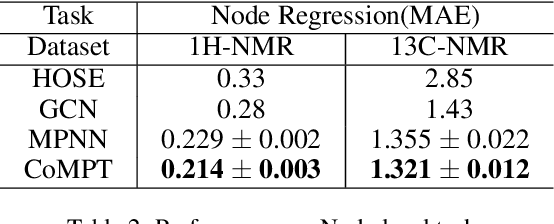
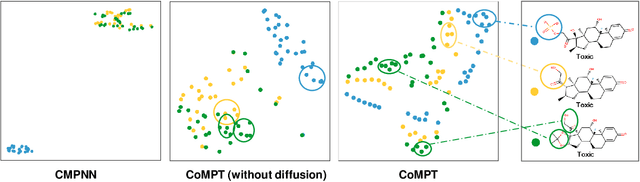
Abstract:Constructing appropriate representations of molecules lies at the core of numerous tasks such as material science, chemistry and drug designs. Recent researches abstract molecules as attributed graphs and employ graph neural networks (GNN) for molecular representation learning, which have made remarkable achievements in molecular graph modeling. Albeit powerful, current models either are based on local aggregation operations and thus miss higher-order graph properties or focus on only node information without fully using the edge information. For this sake, we propose a Communicative Message Passing Transformer (CoMPT) neural network to improve the molecular graph representation by reinforcing message interactions between nodes and edges based on the Transformer architecture. Unlike the previous transformer-style GNNs that treat molecules as fully connected graphs, we introduce a message diffusion mechanism to leverage the graph connectivity inductive bias and reduce the message enrichment explosion. Extensive experiments demonstrated that the proposed model obtained superior performances (around 4$\%$ on average) against state-of-the-art baselines on seven chemical property datasets (graph-level tasks) and two chemical shift datasets (node-level tasks). Further visualization studies also indicated a better representation capacity achieved by our model.
Quantitative Evaluation of Explainable Graph Neural Networks for Molecular Property Prediction
Jul 12, 2021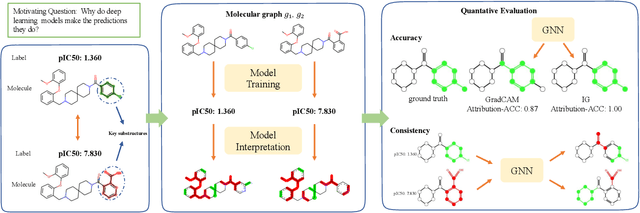
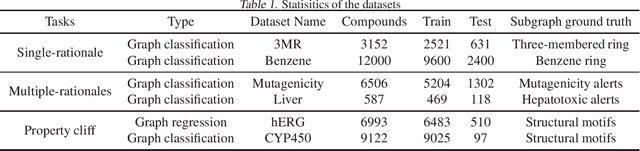
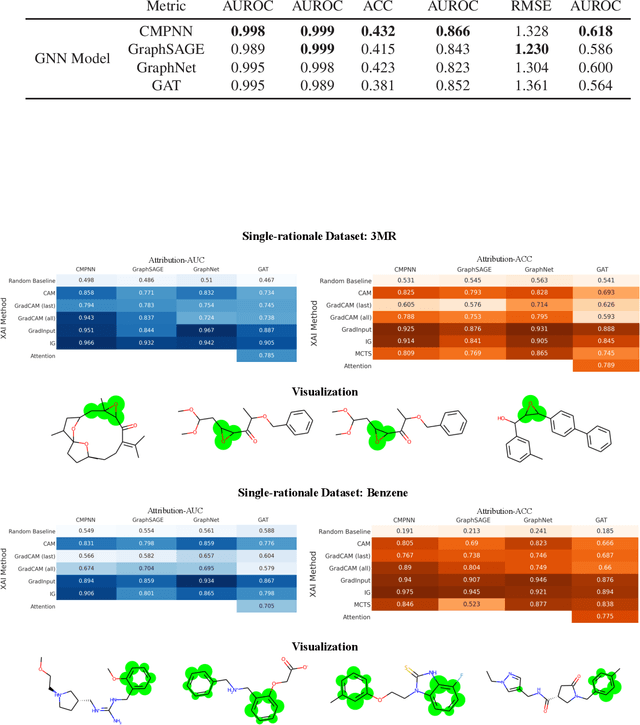
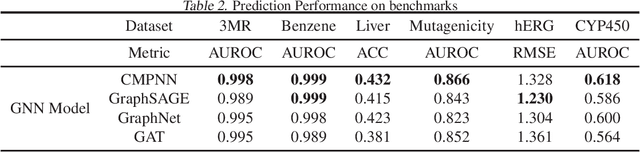
Abstract:Advances in machine learning have led to graph neural network-based methods for drug discovery, yielding promising results in molecular design, chemical synthesis planning, and molecular property prediction. However, current graph neural networks (GNNs) remain of limited acceptance in drug discovery is limited due to their lack of interpretability. Although this major weakness has been mitigated by the development of explainable artificial intelligence (XAI) techniques, the "ground truth" assignment in most explainable tasks ultimately rests with subjective judgments by humans so that the quality of model interpretation is hard to evaluate in quantity. In this work, we first build three levels of benchmark datasets to quantitatively assess the interpretability of the state-of-the-art GNN models. Then we implemented recent XAI methods in combination with different GNN algorithms to highlight the benefits, limitations, and future opportunities for drug discovery. As a result, GradInput and IG generally provide the best model interpretability for GNNs, especially when combined with GraphNet and CMPNN. The integrated and developed XAI package is fully open-sourced and can be used by practitioners to train new models on other drug discovery tasks.
Predicting Retrosynthetic Reaction using Self-Corrected Transformer Neural Networks
Jul 03, 2019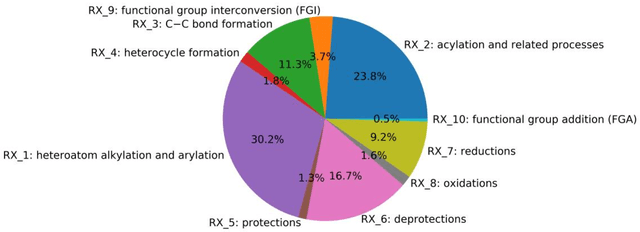
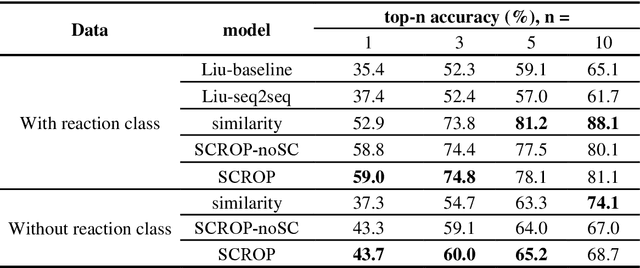
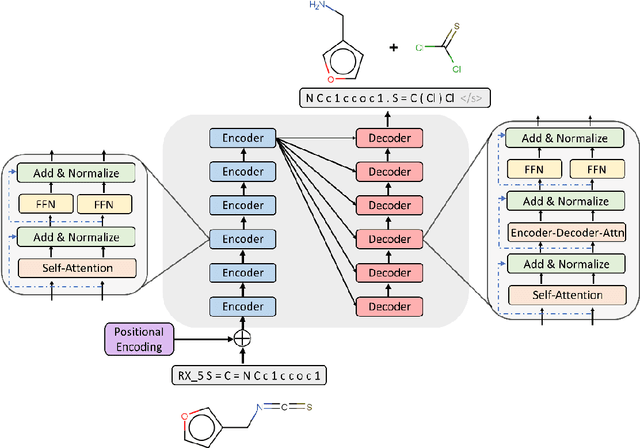
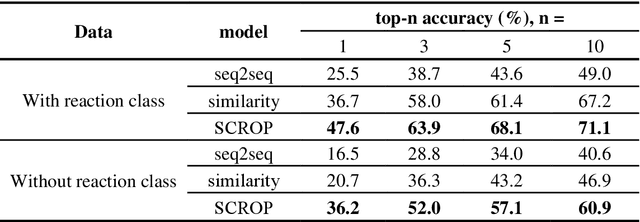
Abstract:Synthesis planning is the process of recursively decomposing target molecules into available precursors. Computer-aided retrosynthesis can potentially assist chemists in designing synthetic routes, but at present it is cumbersome and provides results of dissatisfactory quality. In this study, we develop a template-free self-corrected retrosynthesis predictor (SCROP) to perform a retrosynthesis prediction task trained by using the Transformer neural network architecture. In the method, the retrosynthesis planning is converted as a machine translation problem between molecular linear notations of reactants and the products. Coupled with a neural network-based syntax corrector, our method achieves an accuracy of 59.0% on a standard benchmark dataset, which increases >21% over other deep learning methods, and >6% over template-based methods. More importantly, our method shows an accuracy 1.7 times higher than other state-of-the-art methods for compounds not appearing in the training set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge