Yuedong Yang
De Novo Molecular Generation from Mass Spectra via Many-Body Enhanced Diffusion
Feb 02, 2026Abstract:Molecular structure generation from mass spectrometry is fundamental for understanding cellular metabolism and discovering novel compounds. Although tandem mass spectrometry (MS/MS) enables the high-throughput acquisition of fragment fingerprints, these spectra often reflect higher-order interactions involving the concerted cleavage of multiple atoms and bonds-crucial for resolving complex isomers and non-local fragmentation mechanisms. However, most existing methods adopt atom-centric and pairwise interaction modeling, overlooking higher-order edge interactions and lacking the capacity to systematically capture essential many-body characteristics for structure generation. To overcome these limitations, we present MBGen, a Many-Body enhanced diffusion framework for de novo molecular structure Generation from mass spectra. By integrating a many-body attention mechanism and higher-order edge modeling, MBGen comprehensively leverages the rich structural information encoded in MS/MS spectra, enabling accurate de novo generation and isomer differentiation for novel molecules. Experimental results on the NPLIB1 and MassSpecGym benchmarks demonstrate that MBGen achieves superior performance, with improvements of up to 230% over state-of-the-art methods, highlighting the scientific value and practical utility of many-body modeling for mass spectrometry-based molecular generation. Further analysis and ablation studies show that our approach effectively captures higher-order interactions and exhibits enhanced sensitivity to complex isomeric and non-local fragmentation information.
A 3D pocket-aware and affinity-guided diffusion model for lead optimization
Apr 29, 2025Abstract:Molecular optimization, aimed at improving binding affinity or other molecular properties, is a crucial task in drug discovery that often relies on the expertise of medicinal chemists. Recently, deep learning-based 3D generative models showed promise in enhancing the efficiency of molecular optimization. However, these models often struggle to adequately consider binding affinities with protein targets during lead optimization. Herein, we propose a 3D pocket-aware and affinity-guided diffusion model, named Diffleop, to optimize molecules with enhanced binding affinity. The model explicitly incorporates the knowledge of protein-ligand binding affinity to guide the denoising sampling for molecule generation with high affinity. The comprehensive evaluations indicated that Diffleop outperforms baseline models across multiple metrics, especially in terms of binding affinity.
Strategic priorities for transformative progress in advancing biology with proteomics and artificial intelligence
Feb 21, 2025

Abstract:Artificial intelligence (AI) is transforming scientific research, including proteomics. Advances in mass spectrometry (MS)-based proteomics data quality, diversity, and scale, combined with groundbreaking AI techniques, are unlocking new challenges and opportunities in biological discovery. Here, we highlight key areas where AI is driving innovation, from data analysis to new biological insights. These include developing an AI-friendly ecosystem for proteomics data generation, sharing, and analysis; improving peptide and protein identification and quantification; characterizing protein-protein interactions and protein complexes; advancing spatial and perturbation proteomics; integrating multi-omics data; and ultimately enabling AI-empowered virtual cells.
Incorporating Retrieval-based Causal Learning with Information Bottlenecks for Interpretable Graph Neural Networks
Feb 07, 2024



Abstract:Graph Neural Networks (GNNs) have gained considerable traction for their capability to effectively process topological data, yet their interpretability remains a critical concern. Current interpretation methods are dominated by post-hoc explanations to provide a transparent and intuitive understanding of GNNs. However, they have limited performance in interpreting complicated subgraphs and can't utilize the explanation to advance GNN predictions. On the other hand, transparent GNN models are proposed to capture critical subgraphs. While such methods could improve GNN predictions, they usually don't perform well on explanations. Thus, it is desired for a new strategy to better couple GNN explanation and prediction. In this study, we have developed a novel interpretable causal GNN framework that incorporates retrieval-based causal learning with Graph Information Bottleneck (GIB) theory. The framework could semi-parametrically retrieve crucial subgraphs detected by GIB and compress the explanatory subgraphs via a causal module. The framework was demonstrated to consistently outperform state-of-the-art methods, and to achieve 32.71\% higher precision on real-world explanation scenarios with diverse explanation types. More importantly, the learned explanations were shown able to also improve GNN prediction performance.
Node-based Knowledge Graph Contrastive Learning for Medical Relationship Prediction
Oct 16, 2023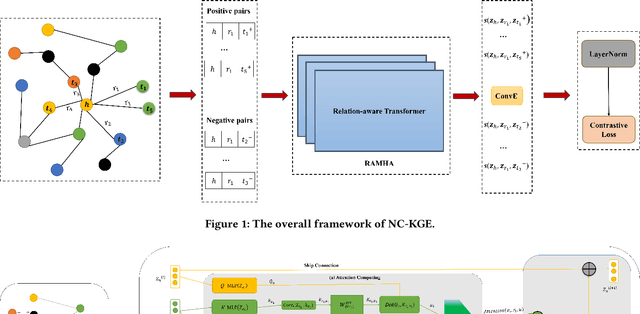

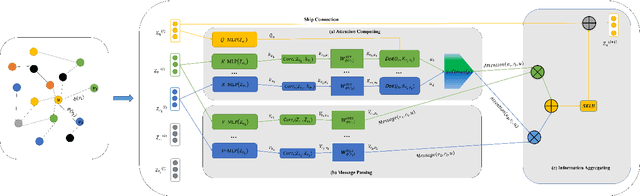

Abstract:The embedding of Biomedical Knowledge Graphs (BKGs) generates robust representations, valuable for a variety of artificial intelligence applications, including predicting drug combinations and reasoning disease-drug relationships. Meanwhile, contrastive learning (CL) is widely employed to enhance the distinctiveness of these representations. However, constructing suitable contrastive pairs for CL, especially within Knowledge Graphs (KGs), has been challenging. In this paper, we proposed a novel node-based contrastive learning method for knowledge graph embedding, NC-KGE. NC-KGE enhances knowledge extraction in embeddings and speeds up training convergence by constructing appropriate contrastive node pairs on KGs. This scheme can be easily integrated with other knowledge graph embedding (KGE) methods. For downstream task such as biochemical relationship prediction, we have incorporated a relation-aware attention mechanism into NC-KGE, focusing on the semantic relationships and node interactions. Extensive experiments show that NC-KGE performs competitively with state-of-the-art models on public datasets like FB15k-237 and WN18RR. Particularly in biomedical relationship prediction tasks, NC-KGE outperforms all baselines on datasets such as PharmKG8k-28, DRKG17k-21, and BioKG72k-14, especially in predicting drug combination relationships. We release our code at https://github.com/zhi520/NC-KGE.
Efficient Low-rank Backpropagation for Vision Transformer Adaptation
Sep 26, 2023Abstract:The increasing scale of vision transformers (ViT) has made the efficient fine-tuning of these large models for specific needs a significant challenge in various applications. This issue originates from the computationally demanding matrix multiplications required during the backpropagation process through linear layers in ViT. In this paper, we tackle this problem by proposing a new Low-rank BackPropagation via Walsh-Hadamard Transformation (LBP-WHT) method. Intuitively, LBP-WHT projects the gradient into a low-rank space and carries out backpropagation. This approach substantially reduces the computation needed for adapting ViT, as matrix multiplication in the low-rank space is far less resource-intensive. We conduct extensive experiments with different models (ViT, hybrid convolution-ViT model) on multiple datasets to demonstrate the effectiveness of our method. For instance, when adapting an EfficientFormer-L1 model on CIFAR100, our LBP-WHT achieves 10.4% higher accuracy than the state-of-the-art baseline, while requiring 9 MFLOPs less computation. As the first work to accelerate ViT adaptation with low-rank backpropagation, our LBP-WHT method is complementary to many prior efforts and can be combined with them for better performance.
TIPS: Topologically Important Path Sampling for Anytime Neural Networks
May 13, 2023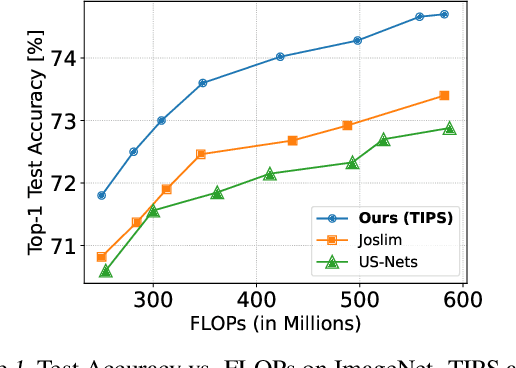
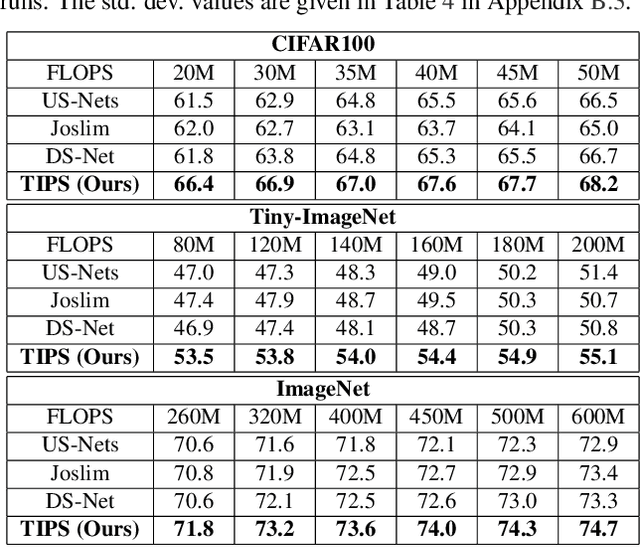
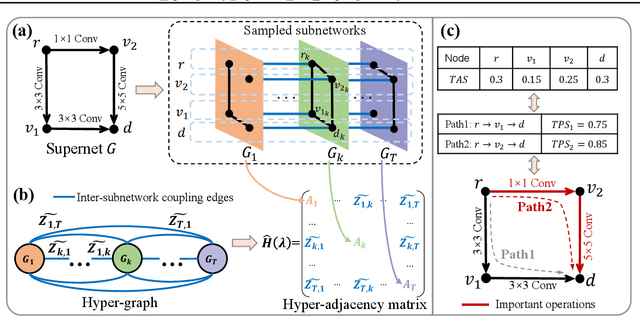
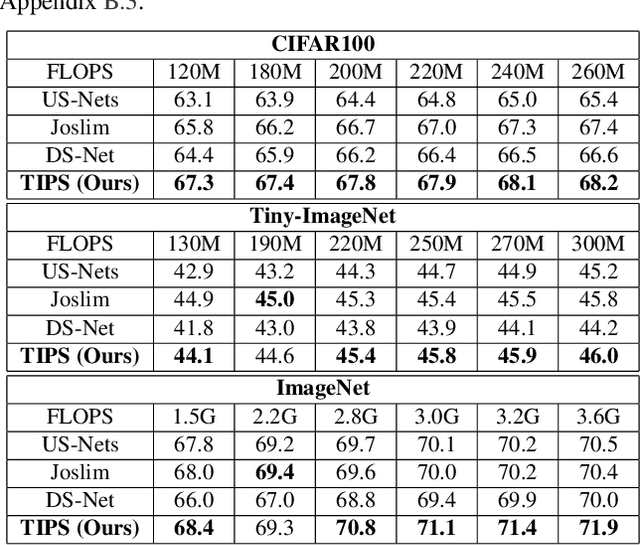
Abstract:Anytime neural networks (AnytimeNNs) are a promising solution to adaptively adjust the model complexity at runtime under various hardware resource constraints. However, the manually-designed AnytimeNNs are biased by designers' prior experience and thus provide sub-optimal solutions. To address the limitations of existing hand-crafted approaches, we first model the training process of AnytimeNNs as a discrete-time Markov chain (DTMC) and use it to identify the paths that contribute the most to the training of AnytimeNNs. Based on this new DTMC-based analysis, we further propose TIPS, a framework to automatically design AnytimeNNs under various hardware constraints. Our experimental results show that TIPS can improve the convergence rate and test accuracy of AnytimeNNs. Compared to the existing AnytimeNNs approaches, TIPS improves the accuracy by 2%-6.6% on multiple datasets and achieves SOTA accuracy-FLOPs tradeoffs.
Retrieval-based Knowledge Augmented Vision Language Pre-training
Apr 27, 2023Abstract:With recent progress in large-scale vision and language representation learning, Vision Language Pretraining (VLP) models have achieved promising improvements on various multi-modal downstream tasks. Albeit powerful, these pre-training models still do not take advantage of world knowledge, which is implicit in multi-modal data but comprises abundant and complementary information. In this work, we propose a REtrieval-based knowledge Augmented Vision Language Pre-training model (REAVL), which retrieves world knowledge from knowledge graphs (KGs) and incorporates them in vision-language pre-training. REAVL has two core components: a knowledge retriever that retrieves knowledge given multi-modal data, and a knowledge-augmented model that fuses multi-modal data and knowledge. By novelly unifying four knowledge-aware self-supervised tasks, REAVL promotes the mutual integration of multi-modal data and knowledge by fusing explicit knowledge with vision-language pairs for masked multi-modal data modeling and KG relational reasoning. Empirical experiments show that REAVL achieves new state-of-the-art performance uniformly on knowledge-based vision-language understanding and multimodal entity linking tasks, and competitive results on general vision-language tasks while only using 0.2% pre-training data of the best models.
ZiCo: Zero-shot NAS via Inverse Coefficient of Variation on Gradients
Jan 26, 2023



Abstract:Neural Architecture Search (NAS) is widely used to automatically design the neural network with the best performance among a large number of candidate architectures. To reduce the search time, zero-shot NAS aims at designing training-free proxies that can predict the test performance of a given architecture. However, as shown recently, none of the zero-shot proxies proposed to date can actually work consistently better than a naive proxy, namely, the number of network parameters (#Params). To improve this state of affairs, as the main theoretical contribution, we first reveal how some specific gradient properties across different samples impact the convergence rate and generalization capacity of neural networks. Based on this theoretical analysis, we propose a new zero-shot proxy, ZiCo, the first proxy that works consistently better than #Params. We demonstrate that ZiCo works better than State-Of-The-Art (SOTA) proxies on several popular NAS-Benchmarks (NASBench101, NATSBench-SSS/TSS, TransNASBench-101) for multiple applications (e.g., image classification/reconstruction and pixel-level prediction). Finally, we demonstrate that the optimal architectures found via ZiCo are as competitive as the ones found by one-shot and multi-shot NAS methods, but with much less search time. For example, ZiCo-based NAS can find optimal architectures with 78.1%, 79.4%, and 80.4% test accuracy under inference budgets of 450M, 600M, and 1000M FLOPs on ImageNet within 0.4 GPU days.
Efficient On-device Training via Gradient Filtering
Jan 01, 2023Abstract:Despite its importance for federated learning, continuous learning and many other applications, on-device training remains an open problem for EdgeAI. The problem stems from the large number of operations (e.g., floating point multiplications and additions) and memory consumption required during training by the back-propagation algorithm. Consequently, in this paper, we propose a new gradient filtering approach which enables on-device DNN model training. More precisely, our approach creates a special structure with fewer unique elements in the gradient map, thus significantly reducing the computational complexity and memory consumption of back propagation during training. Extensive experiments on image classification and semantic segmentation with multiple DNN models (e.g., MobileNet, DeepLabV3, UPerNet) and devices (e.g., Raspberry Pi and Jetson Nano) demonstrate the effectiveness and wide applicability of our approach. For example, compared to SOTA, we achieve up to 19$\times$ speedup and 77.1% memory savings on ImageNet classification with only 0.1% accuracy loss. Finally, our method is easy to implement and deploy; over 20$\times$ speedup and 90% energy savings have been observed compared to highly optimized baselines in MKLDNN and CUDNN on NVIDIA Jetson Nano. Consequently, our approach opens up a new direction of research with a huge potential for on-device training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge