Node-based Knowledge Graph Contrastive Learning for Medical Relationship Prediction
Paper and Code
Oct 16, 2023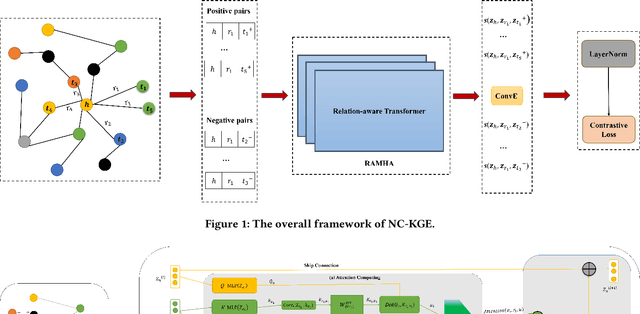

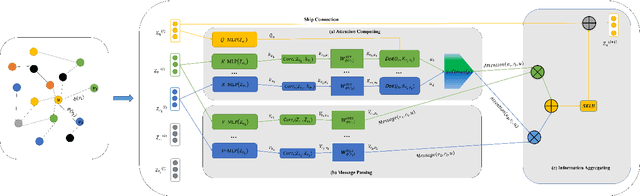

The embedding of Biomedical Knowledge Graphs (BKGs) generates robust representations, valuable for a variety of artificial intelligence applications, including predicting drug combinations and reasoning disease-drug relationships. Meanwhile, contrastive learning (CL) is widely employed to enhance the distinctiveness of these representations. However, constructing suitable contrastive pairs for CL, especially within Knowledge Graphs (KGs), has been challenging. In this paper, we proposed a novel node-based contrastive learning method for knowledge graph embedding, NC-KGE. NC-KGE enhances knowledge extraction in embeddings and speeds up training convergence by constructing appropriate contrastive node pairs on KGs. This scheme can be easily integrated with other knowledge graph embedding (KGE) methods. For downstream task such as biochemical relationship prediction, we have incorporated a relation-aware attention mechanism into NC-KGE, focusing on the semantic relationships and node interactions. Extensive experiments show that NC-KGE performs competitively with state-of-the-art models on public datasets like FB15k-237 and WN18RR. Particularly in biomedical relationship prediction tasks, NC-KGE outperforms all baselines on datasets such as PharmKG8k-28, DRKG17k-21, and BioKG72k-14, especially in predicting drug combination relationships. We release our code at https://github.com/zhi520/NC-KGE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge