Edmond Boyer
Combining Neural Fields and Deformation Models for Non-Rigid 3D Motion Reconstruction from Partial Data
Dec 11, 2024



Abstract:We introduce a novel, data-driven approach for reconstructing temporally coherent 3D motion from unstructured and potentially partial observations of non-rigidly deforming shapes. Our goal is to achieve high-fidelity motion reconstructions for shapes that undergo near-isometric deformations, such as humans wearing loose clothing. The key novelty of our work lies in its ability to combine implicit shape representations with explicit mesh-based deformation models, enabling detailed and temporally coherent motion reconstructions without relying on parametric shape models or decoupling shape and motion. Each frame is represented as a neural field decoded from a feature space where observations over time are fused, hence preserving geometric details present in the input data. Temporal coherence is enforced with a near-isometric deformation constraint between adjacent frames that applies to the underlying surface in the neural field. Our method outperforms state-of-the-art approaches, as demonstrated by its application to human and animal motion sequences reconstructed from monocular depth videos.
SplatFields: Neural Gaussian Splats for Sparse 3D and 4D Reconstruction
Sep 17, 2024



Abstract:Digitizing 3D static scenes and 4D dynamic events from multi-view images has long been a challenge in computer vision and graphics. Recently, 3D Gaussian Splatting (3DGS) has emerged as a practical and scalable reconstruction method, gaining popularity due to its impressive reconstruction quality, real-time rendering capabilities, and compatibility with widely used visualization tools. However, the method requires a substantial number of input views to achieve high-quality scene reconstruction, introducing a significant practical bottleneck. This challenge is especially severe in capturing dynamic scenes, where deploying an extensive camera array can be prohibitively costly. In this work, we identify the lack of spatial autocorrelation of splat features as one of the factors contributing to the suboptimal performance of the 3DGS technique in sparse reconstruction settings. To address the issue, we propose an optimization strategy that effectively regularizes splat features by modeling them as the outputs of a corresponding implicit neural field. This results in a consistent enhancement of reconstruction quality across various scenarios. Our approach effectively handles static and dynamic cases, as demonstrated by extensive testing across different setups and scene complexities.
VortSDF: 3D Modeling with Centroidal Voronoi Tesselation on Signed Distance Field
Jul 29, 2024Abstract:Volumetric shape representations have become ubiquitous in multi-view reconstruction tasks. They often build on regular voxel grids as discrete representations of 3D shape functions, such as SDF or radiance fields, either as the full shape model or as sampled instantiations of continuous representations, as with neural networks. Despite their proven efficiency, voxel representations come with the precision versus complexity trade-off. This inherent limitation can significantly impact performance when moving away from simple and uncluttered scenes. In this paper we investigate an alternative discretization strategy with the Centroidal Voronoi Tesselation (CVT). CVTs allow to better partition the observation space with respect to shape occupancy and to focus the discretization around shape surfaces. To leverage this discretization strategy for multi-view reconstruction, we introduce a volumetric optimization framework that combines explicit SDF fields with a shallow color network, in order to estimate 3D shape properties over tetrahedral grids. Experimental results with Chamfer statistics validate this approach with unprecedented reconstruction quality on various scenarios such as objects, open scenes or human.
ANIM: Accurate Neural Implicit Model for Human Reconstruction from a single RGB-D image
Mar 18, 2024



Abstract:Recent progress in human shape learning, shows that neural implicit models are effective in generating 3D human surfaces from limited number of views, and even from a single RGB image. However, existing monocular approaches still struggle to recover fine geometric details such as face, hands or cloth wrinkles. They are also easily prone to depth ambiguities that result in distorted geometries along the camera optical axis. In this paper, we explore the benefits of incorporating depth observations in the reconstruction process by introducing ANIM, a novel method that reconstructs arbitrary 3D human shapes from single-view RGB-D images with an unprecedented level of accuracy. Our model learns geometric details from both multi-resolution pixel-aligned and voxel-aligned features to leverage depth information and enable spatial relationships, mitigating depth ambiguities. We further enhance the quality of the reconstructed shape by introducing a depth-supervision strategy, which improves the accuracy of the signed distance field estimation of points that lie on the reconstructed surface. Experiments demonstrate that ANIM outperforms state-of-the-art works that use RGB, surface normals, point cloud or RGB-D data as input. In addition, we introduce ANIM-Real, a new multi-modal dataset comprising high-quality scans paired with consumer-grade RGB-D camera, and our protocol to fine-tune ANIM, enabling high-quality reconstruction from real-world human capture.
HISR: Hybrid Implicit Surface Representation for Photorealistic 3D Human Reconstruction
Dec 28, 2023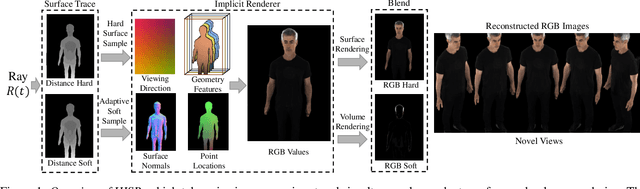
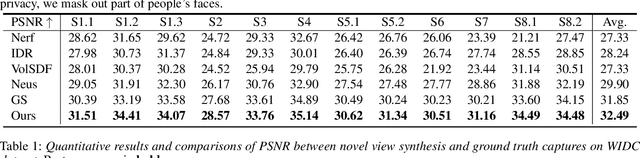
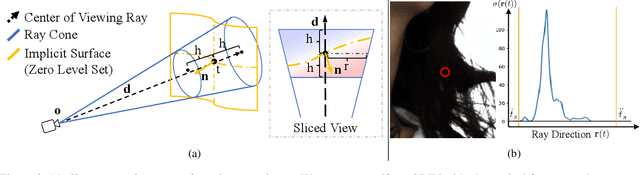
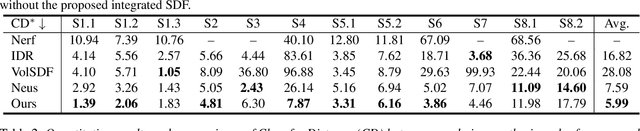
Abstract:Neural reconstruction and rendering strategies have demonstrated state-of-the-art performances due, in part, to their ability to preserve high level shape details. Existing approaches, however, either represent objects as implicit surface functions or neural volumes and still struggle to recover shapes with heterogeneous materials, in particular human skin, hair or clothes. To this aim, we present a new hybrid implicit surface representation to model human shapes. This representation is composed of two surface layers that represent opaque and translucent regions on the clothed human body. We segment different regions automatically using visual cues and learn to reconstruct two signed distance functions (SDFs). We perform surface-based rendering on opaque regions (e.g., body, face, clothes) to preserve high-fidelity surface normals and volume rendering on translucent regions (e.g., hair). Experiments demonstrate that our approach obtains state-of-the-art results on 3D human reconstructions, and also shows competitive performances on other objects.
Deformation-Guided Unsupervised Non-Rigid Shape Matching
Nov 27, 2023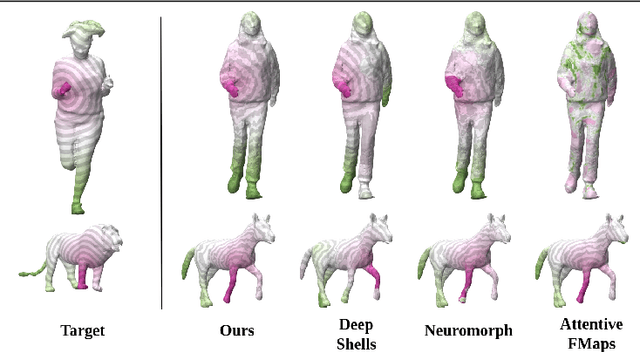
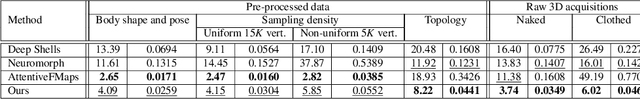
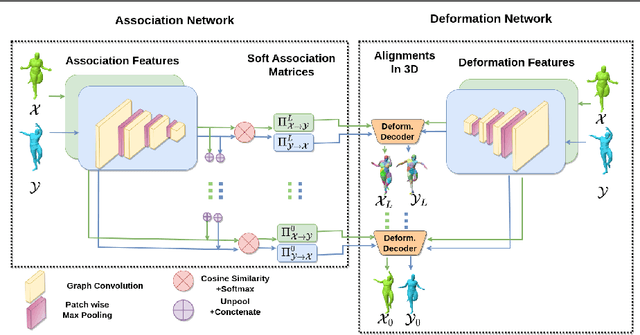
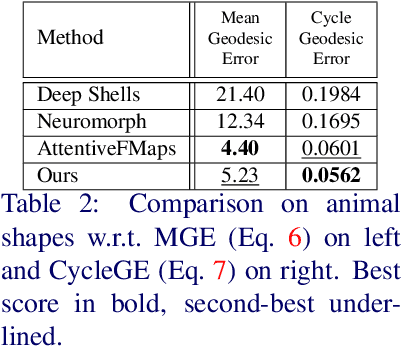
Abstract:We present an unsupervised data-driven approach for non-rigid shape matching. Shape matching identifies correspondences between two shapes and is a fundamental step in many computer vision and graphics applications. Our approach is designed to be particularly robust when matching shapes digitized using 3D scanners that contain fine geometric detail and suffer from different types of noise including topological noise caused by the coalescence of spatially close surface regions. We build on two strategies. First, using a hierarchical patch based shape representation we match shapes consistently in a coarse to fine manner, allowing for robustness to noise. This multi-scale representation drastically reduces the dimensionality of the problem when matching at the coarsest scale, rendering unsupervised learning feasible. Second, we constrain this hierarchical matching to be reflected in 3D by fitting a patch-wise near-rigid deformation model. Using this constraint, we leverage spatial continuity at different scales to capture global shape properties, resulting in matchings that generalize well to data with different deformations and noise characteristics. Experiments demonstrate that our approach obtains significantly better results on raw 3D scans than state-of-the-art methods, while performing on-par on standard test scenarios.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
4DHumanOutfit: a multi-subject 4D dataset of human motion sequences in varying outfits exhibiting large displacements
Jun 12, 2023
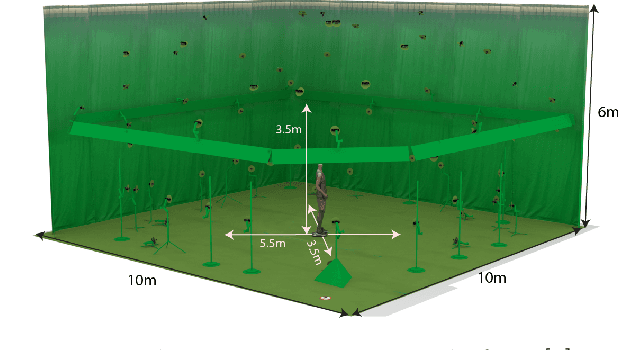

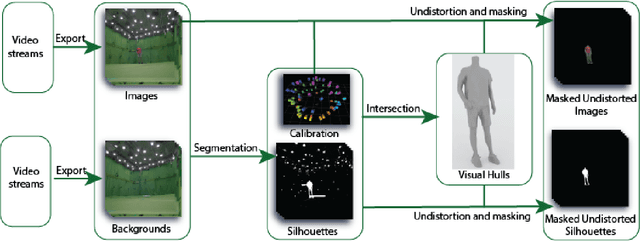
Abstract:This work presents 4DHumanOutfit, a new dataset of densely sampled spatio-temporal 4D human motion data of different actors, outfits and motions. The dataset is designed to contain different actors wearing different outfits while performing different motions in each outfit. In this way, the dataset can be seen as a cube of data containing 4D motion sequences along 3 axes with identity, outfit and motion. This rich dataset has numerous potential applications for the processing and creation of digital humans, e.g. augmented reality, avatar creation and virtual try on. 4DHumanOutfit is released for research purposes at https://kinovis.inria.fr/4dhumanoutfit/. In addition to image data and 4D reconstructions, the dataset includes reference solutions for each axis. We present independent baselines along each axis that demonstrate the value of these reference solutions for evaluation tasks.
3DTeethSeg'22: 3D Teeth Scan Segmentation and Labeling Challenge
May 29, 2023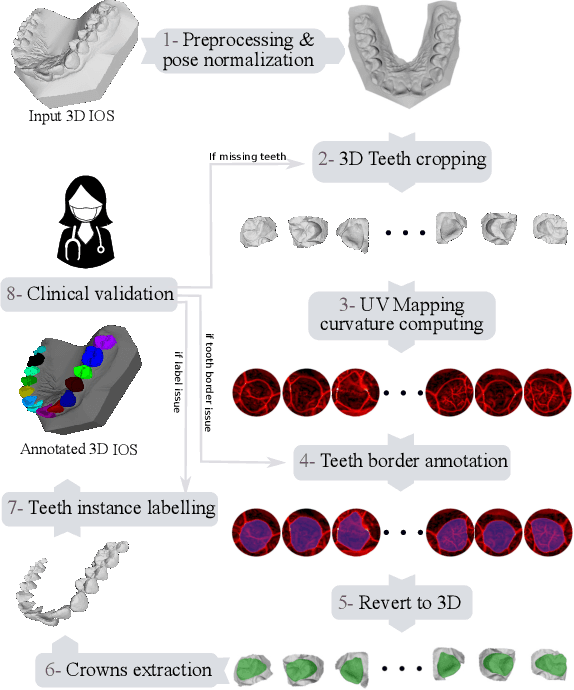
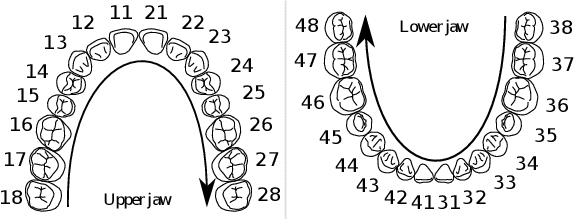
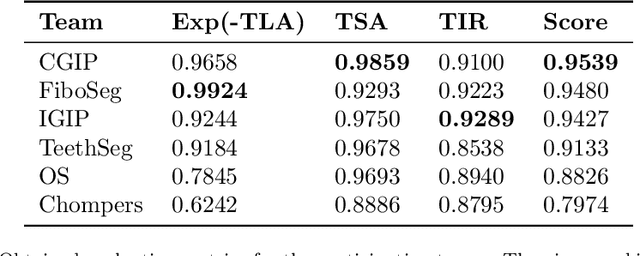
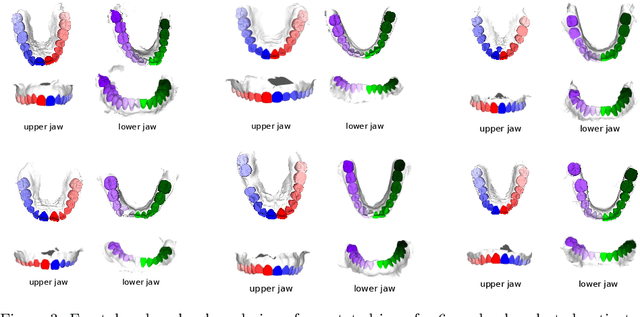
Abstract:Teeth localization, segmentation, and labeling from intra-oral 3D scans are essential tasks in modern dentistry to enhance dental diagnostics, treatment planning, and population-based studies on oral health. However, developing automated algorithms for teeth analysis presents significant challenges due to variations in dental anatomy, imaging protocols, and limited availability of publicly accessible data. To address these challenges, the 3DTeethSeg'22 challenge was organized in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) in 2022, with a call for algorithms tackling teeth localization, segmentation, and labeling from intraoral 3D scans. A dataset comprising a total of 1800 scans from 900 patients was prepared, and each tooth was individually annotated by a human-machine hybrid algorithm. A total of 6 algorithms were evaluated on this dataset. In this study, we present the evaluation results of the 3DTeethSeg'22 challenge. The 3DTeethSeg'22 challenge code can be accessed at: https://github.com/abenhamadou/3DTeethSeg22_challenge
Teeth3DS: a benchmark for teeth segmentation and labeling from intra-oral 3D scans
Oct 12, 2022



Abstract:Teeth segmentation and labeling are critical components of Computer-Aided Dentistry (CAD) systems. Indeed, before any orthodontic or prosthetic treatment planning, a CAD system needs to first accurately segment and label each instance of teeth visible in the 3D dental scan, this is to avoid time-consuming manual adjustments by the dentist. Nevertheless, developing such an automated and accurate dental segmentation and labeling tool is very challenging, especially given the lack of publicly available datasets or benchmarks. This article introduces the first public benchmark, named Teeth3DS, which has been created in the frame of the 3DTeethSeg 2022 MICCAI challenge to boost the research field and inspire the 3D vision research community to work on intra-oral 3D scans analysis such as teeth identification, segmentation, labeling, 3D modeling and 3D reconstruction. Teeth3DS is made of 1800 intra-oral scans (23999 annotated teeth) collected from 900 patients covering the upper and lower jaws separately, acquired and validated by orthodontists/dental surgeons with more than 5 years of professional experience.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge