Achraf Ben-Hamadou
Detecting Dental Landmarks from Intraoral 3D Scans: the 3DTeethLand challenge
Dec 09, 2025Abstract:Teeth landmark detection is a critical task in modern clinical orthodontics. Their precise identification enables advanced diagnostics, facilitates personalized treatment strategies, and supports more effective monitoring of treatment progress in clinical dentistry. However, several significant challenges may arise due to the intricate geometry of individual teeth and the substantial variations observed across different individuals. To address these complexities, the development of advanced techniques, especially through the application of deep learning, is essential for the precise and reliable detection of 3D tooth landmarks. In this context, the 3DTeethLand challenge was held in collaboration with the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) in 2024, calling for algorithms focused on teeth landmark detection from intraoral 3D scans. This challenge introduced the first publicly available dataset for 3D teeth landmark detection, offering a valuable resource to assess the state-of-the-art methods in this task and encourage the community to provide methodological contributions towards the resolution of their problem with significant clinical implications.
Cross-Attention Fusion of Visual and Geometric Features for Large Vocabulary Arabic Lipreading
Feb 18, 2024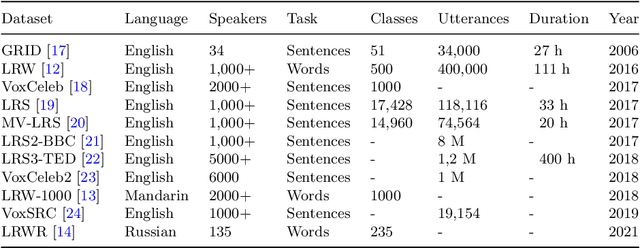



Abstract:Lipreading involves using visual data to recognize spoken words by analyzing the movements of the lips and surrounding area. It is a hot research topic with many potential applications, such as human-machine interaction and enhancing audio speech recognition. Recent deep-learning based works aim to integrate visual features extracted from the mouth region with landmark points on the lip contours. However, employing a simple combination method such as concatenation may not be the most effective approach to get the optimal feature vector. To address this challenge, firstly, we propose a cross-attention fusion-based approach for large lexicon Arabic vocabulary to predict spoken words in videos. Our method leverages the power of cross-attention networks to efficiently integrate visual and geometric features computed on the mouth region. Secondly, we introduce the first large-scale Lip Reading in the Wild for Arabic (LRW-AR) dataset containing 20,000 videos for 100-word classes, uttered by 36 speakers. The experimental results obtained on LRW-AR and ArabicVisual databases showed the effectiveness and robustness of the proposed approach in recognizing Arabic words. Our work provides insights into the feasibility and effectiveness of applying lipreading techniques to the Arabic language, opening doors for further research in this field. Link to the project page: https://crns-smartvision.github.io/lrwar
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
Deep Generative Models for Physiological Signals: A Systematic Literature Review
Jul 12, 2023Abstract:In this paper, we present a systematic literature review on deep generative models for physiological signals, particularly electrocardiogram, electroencephalogram, photoplethysmogram and electromyogram. Compared to the existing review papers, we present the first review that summarizes the recent state-of-the-art deep generative models. By analysing the state-of-the-art research related to deep generative models along with their main applications and challenges, this review contributes to the overall understanding of these models applied to physiological signals. Additionally, by highlighting the employed evaluation protocol and the most used physiological databases, this review facilitates the assessment and benchmarking of deep generative models.
Graph Self-Supervised Learning for Endoscopic Image Matching
Jun 19, 2023Abstract:Accurate feature matching and correspondence in endoscopic images play a crucial role in various clinical applications, including patient follow-up and rapid anomaly localization through panoramic image generation. However, developing robust and accurate feature matching techniques faces challenges due to the lack of discriminative texture and significant variability between patients. To address these limitations, we propose a novel self-supervised approach that combines Convolutional Neural Networks for capturing local visual appearance and attention-based Graph Neural Networks for modeling spatial relationships between key-points. Our approach is trained in a fully self-supervised scheme without the need for labeled data. Our approach outperforms state-of-the-art handcrafted and deep learning-based methods, demonstrating exceptional performance in terms of precision rate (1) and matching score (99.3%). We also provide code and materials related to this work, which can be accessed at https://github.com/abenhamadou/graph-self-supervised-learning-for-endoscopic-image-matching.
DiffECG: A Generalized Probabilistic Diffusion Model for ECG Signals Synthesis
Jun 02, 2023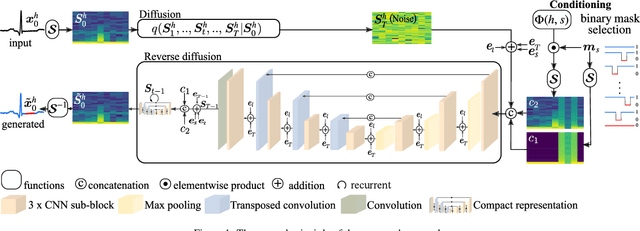
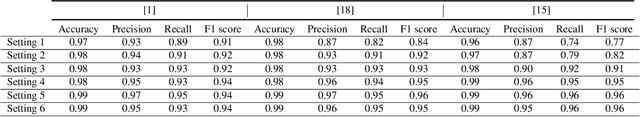
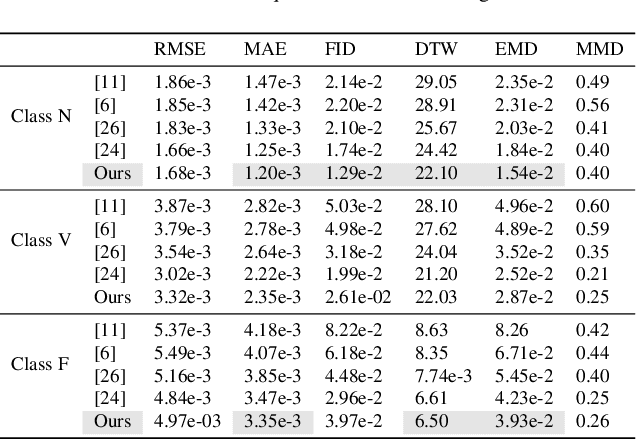
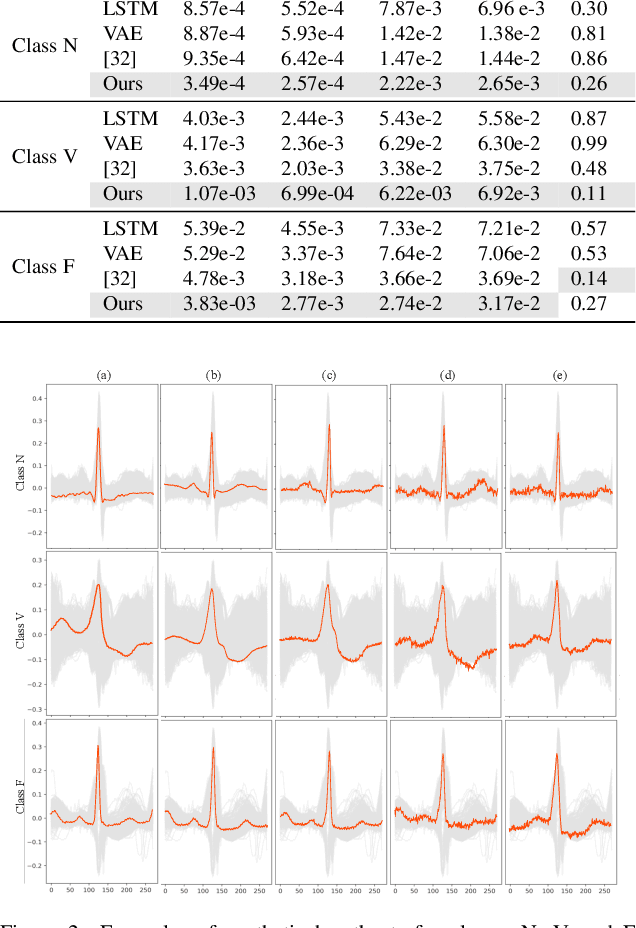
Abstract:In recent years, deep generative models have gained attention as a promising data augmentation solution for heart disease detection using deep learning approaches applied to ECG signals. In this paper, we introduce a novel approach based on denoising diffusion probabilistic models for ECG synthesis that covers three scenarios: heartbeat generation, partial signal completion, and full heartbeat forecasting. Our approach represents the first generalized conditional approach for ECG synthesis, and our experimental results demonstrate its effectiveness for various ECG-related tasks. Moreover, we show that our approach outperforms other state-of-the-art ECG generative models and can enhance the performance of state-of-the-art classifiers.
3DTeethSeg'22: 3D Teeth Scan Segmentation and Labeling Challenge
May 29, 2023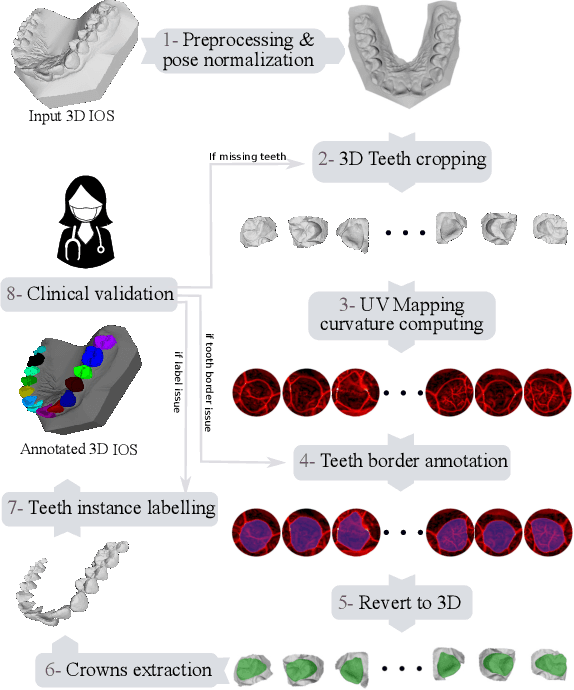
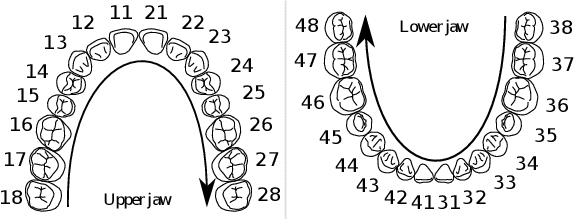
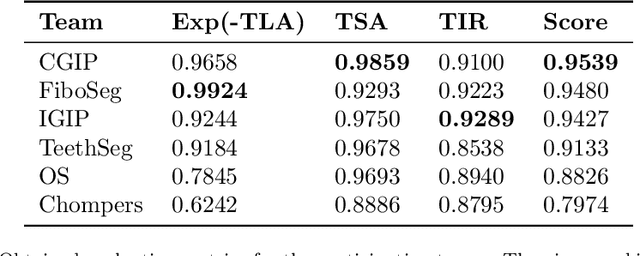
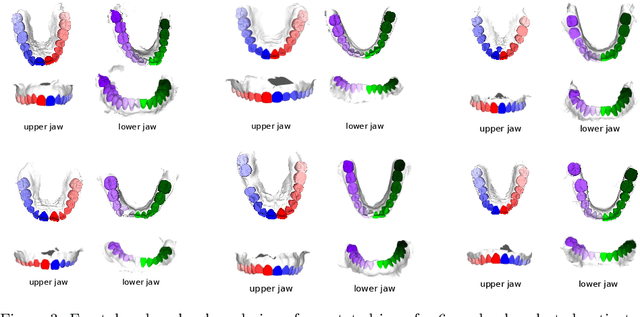
Abstract:Teeth localization, segmentation, and labeling from intra-oral 3D scans are essential tasks in modern dentistry to enhance dental diagnostics, treatment planning, and population-based studies on oral health. However, developing automated algorithms for teeth analysis presents significant challenges due to variations in dental anatomy, imaging protocols, and limited availability of publicly accessible data. To address these challenges, the 3DTeethSeg'22 challenge was organized in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) in 2022, with a call for algorithms tackling teeth localization, segmentation, and labeling from intraoral 3D scans. A dataset comprising a total of 1800 scans from 900 patients was prepared, and each tooth was individually annotated by a human-machine hybrid algorithm. A total of 6 algorithms were evaluated on this dataset. In this study, we present the evaluation results of the 3DTeethSeg'22 challenge. The 3DTeethSeg'22 challenge code can be accessed at: https://github.com/abenhamadou/3DTeethSeg22_challenge
Leveraging Statistical Shape Priors in GAN-based ECG Synthesis
Oct 22, 2022Abstract:Due to the difficulty of collecting electrocardiogram (ECG) data during emergency situations, ECG data generation is an efficient solution for dealing with highly imbalanced ECG training datasets. However, due to the complex dynamics of ECG signals, the synthesis of such signals is a challenging task. In this paper, we present a novel approach for ECG signal generation based on Generative Adversarial Networks (GANs). Our approach combines GANs with statistical ECG data modeling to leverage prior knowledge about ECG dynamics in the generation process. To validate the proposed approach, we present experiments using ECG signals from the MIT-BIH arrhythmia database. The obtained results show the benefits of modeling temporal and amplitude variations of ECG signals as 2-D shapes in generating realistic signals and also improving the performance of state-of-the-art arrhythmia classification baselines.
Teeth3DS: a benchmark for teeth segmentation and labeling from intra-oral 3D scans
Oct 12, 2022



Abstract:Teeth segmentation and labeling are critical components of Computer-Aided Dentistry (CAD) systems. Indeed, before any orthodontic or prosthetic treatment planning, a CAD system needs to first accurately segment and label each instance of teeth visible in the 3D dental scan, this is to avoid time-consuming manual adjustments by the dentist. Nevertheless, developing such an automated and accurate dental segmentation and labeling tool is very challenging, especially given the lack of publicly available datasets or benchmarks. This article introduces the first public benchmark, named Teeth3DS, which has been created in the frame of the 3DTeethSeg 2022 MICCAI challenge to boost the research field and inspire the 3D vision research community to work on intra-oral 3D scans analysis such as teeth identification, segmentation, labeling, 3D modeling and 3D reconstruction. Teeth3DS is made of 1800 intra-oral scans (23999 annotated teeth) collected from 900 patients covering the upper and lower jaws separately, acquired and validated by orthodontists/dental surgeons with more than 5 years of professional experience.
Self-Supervised Endoscopic Image Key-Points Matching
Aug 24, 2022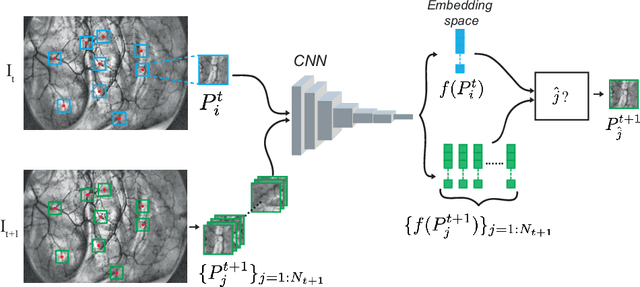
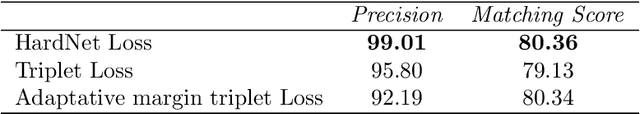
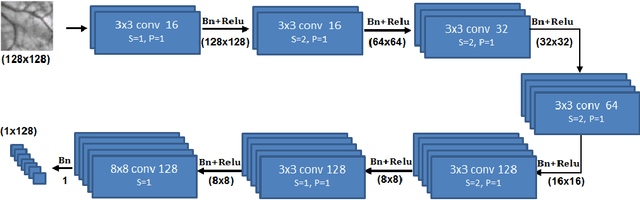
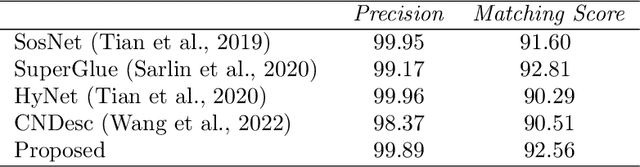
Abstract:Feature matching and finding correspondences between endoscopic images is a key step in many clinical applications such as patient follow-up and generation of panoramic image from clinical sequences for fast anomalies localization. Nonetheless, due to the high texture variability present in endoscopic images, the development of robust and accurate feature matching becomes a challenging task. Recently, deep learning techniques which deliver learned features extracted via convolutional neural networks (CNNs) have gained traction in a wide range of computer vision tasks. However, they all follow a supervised learning scheme where a large amount of annotated data is required to reach good performances, which is generally not always available for medical data databases. To overcome this limitation related to labeled data scarcity, the self-supervised learning paradigm has recently shown great success in a number of applications. This paper proposes a novel self-supervised approach for endoscopic image matching based on deep learning techniques. When compared to standard hand-crafted local feature descriptors, our method outperformed them in terms of precision and recall. Furthermore, our self-supervised descriptor provides a competitive performance in comparison to a selection of state-of-the-art deep learning based supervised methods in terms of precision and matching score.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge