Sergi Pujades
Detecting Dental Landmarks from Intraoral 3D Scans: the 3DTeethLand challenge
Dec 09, 2025Abstract:Teeth landmark detection is a critical task in modern clinical orthodontics. Their precise identification enables advanced diagnostics, facilitates personalized treatment strategies, and supports more effective monitoring of treatment progress in clinical dentistry. However, several significant challenges may arise due to the intricate geometry of individual teeth and the substantial variations observed across different individuals. To address these complexities, the development of advanced techniques, especially through the application of deep learning, is essential for the precise and reliable detection of 3D tooth landmarks. In this context, the 3DTeethLand challenge was held in collaboration with the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) in 2024, calling for algorithms focused on teeth landmark detection from intraoral 3D scans. This challenge introduced the first publicly available dataset for 3D teeth landmark detection, offering a valuable resource to assess the state-of-the-art methods in this task and encourage the community to provide methodological contributions towards the resolution of their problem with significant clinical implications.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
4DHumanOutfit: a multi-subject 4D dataset of human motion sequences in varying outfits exhibiting large displacements
Jun 12, 2023
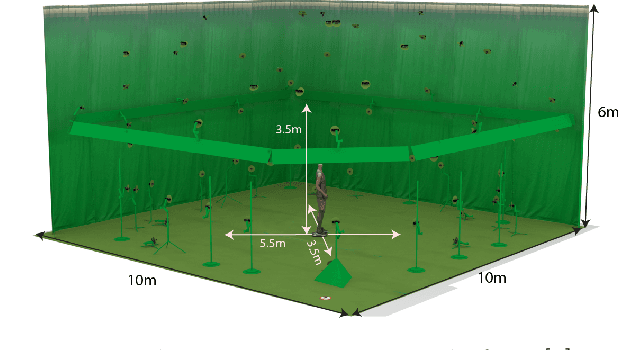

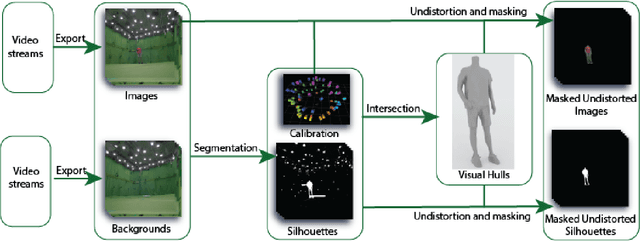
Abstract:This work presents 4DHumanOutfit, a new dataset of densely sampled spatio-temporal 4D human motion data of different actors, outfits and motions. The dataset is designed to contain different actors wearing different outfits while performing different motions in each outfit. In this way, the dataset can be seen as a cube of data containing 4D motion sequences along 3 axes with identity, outfit and motion. This rich dataset has numerous potential applications for the processing and creation of digital humans, e.g. augmented reality, avatar creation and virtual try on. 4DHumanOutfit is released for research purposes at https://kinovis.inria.fr/4dhumanoutfit/. In addition to image data and 4D reconstructions, the dataset includes reference solutions for each axis. We present independent baselines along each axis that demonstrate the value of these reference solutions for evaluation tasks.
3DTeethSeg'22: 3D Teeth Scan Segmentation and Labeling Challenge
May 29, 2023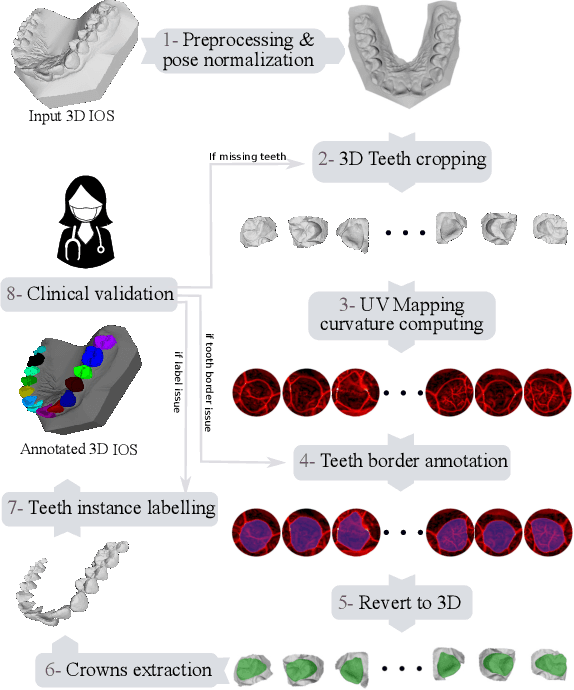
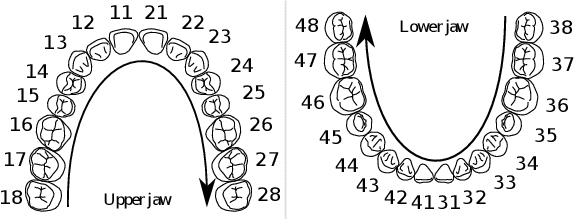
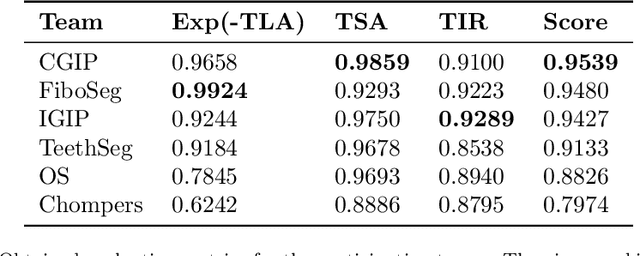
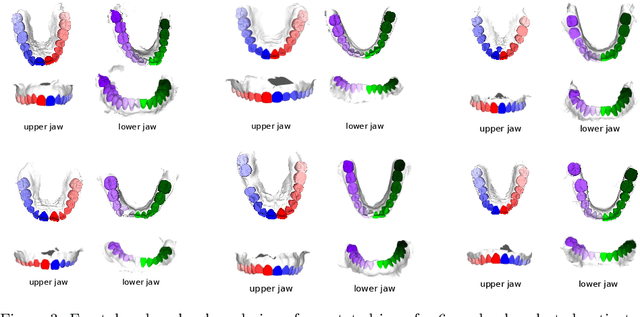
Abstract:Teeth localization, segmentation, and labeling from intra-oral 3D scans are essential tasks in modern dentistry to enhance dental diagnostics, treatment planning, and population-based studies on oral health. However, developing automated algorithms for teeth analysis presents significant challenges due to variations in dental anatomy, imaging protocols, and limited availability of publicly accessible data. To address these challenges, the 3DTeethSeg'22 challenge was organized in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) in 2022, with a call for algorithms tackling teeth localization, segmentation, and labeling from intraoral 3D scans. A dataset comprising a total of 1800 scans from 900 patients was prepared, and each tooth was individually annotated by a human-machine hybrid algorithm. A total of 6 algorithms were evaluated on this dataset. In this study, we present the evaluation results of the 3DTeethSeg'22 challenge. The 3DTeethSeg'22 challenge code can be accessed at: https://github.com/abenhamadou/3DTeethSeg22_challenge
Teeth3DS: a benchmark for teeth segmentation and labeling from intra-oral 3D scans
Oct 12, 2022



Abstract:Teeth segmentation and labeling are critical components of Computer-Aided Dentistry (CAD) systems. Indeed, before any orthodontic or prosthetic treatment planning, a CAD system needs to first accurately segment and label each instance of teeth visible in the 3D dental scan, this is to avoid time-consuming manual adjustments by the dentist. Nevertheless, developing such an automated and accurate dental segmentation and labeling tool is very challenging, especially given the lack of publicly available datasets or benchmarks. This article introduces the first public benchmark, named Teeth3DS, which has been created in the frame of the 3DTeethSeg 2022 MICCAI challenge to boost the research field and inspire the 3D vision research community to work on intra-oral 3D scans analysis such as teeth identification, segmentation, labeling, 3D modeling and 3D reconstruction. Teeth3DS is made of 1800 intra-oral scans (23999 annotated teeth) collected from 900 patients covering the upper and lower jaws separately, acquired and validated by orthodontists/dental surgeons with more than 5 years of professional experience.
OSSO: Obtaining Skeletal Shape from Outside
Apr 21, 2022

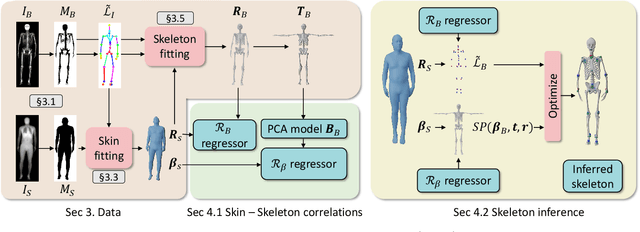
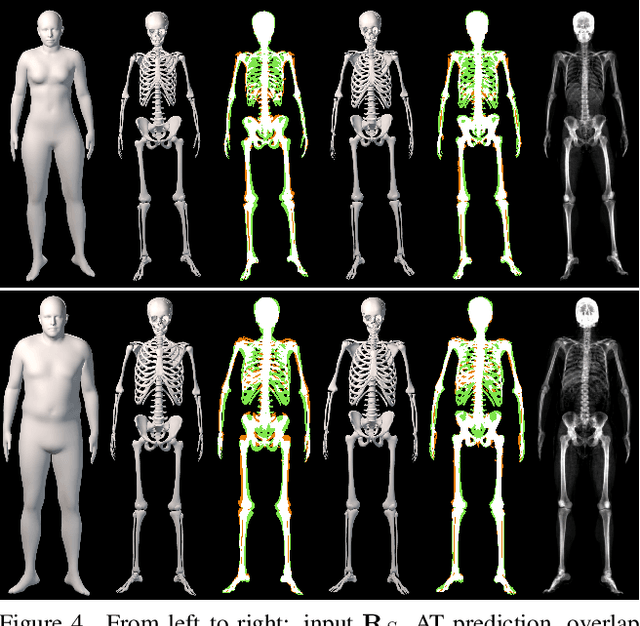
Abstract:We address the problem of inferring the anatomic skeleton of a person, in an arbitrary pose, from the 3D surface of the body; i.e. we predict the inside (bones) from the outside (skin). This has many applications in medicine and biomechanics. Existing state-of-the-art biomechanical skeletons are detailed but do not easily generalize to new subjects. Additionally, computer vision and graphics methods that predict skeletons are typically heuristic, not learned from data, do not leverage the full 3D body surface, and are not validated against ground truth. To our knowledge, our system, called OSSO (Obtaining Skeletal Shape from Outside), is the first to learn the mapping from the 3D body surface to the internal skeleton from real data. We do so using 1000 male and 1000 female dual-energy X-ray absorptiometry (DXA) scans. To these, we fit a parametric 3D body shape model (STAR) to capture the body surface and a novel part-based 3D skeleton model to capture the bones. This provides inside/outside training pairs. We model the statistical variation of full skeletons using PCA in a pose-normalized space. We then train a regressor from body shape parameters to skeleton shape parameters and refine the skeleton to satisfy constraints on physical plausibility. Given an arbitrary 3D body shape and pose, OSSO predicts a realistic skeleton inside. In contrast to previous work, we evaluate the accuracy of the skeleton shape quantitatively on held-out DXA scans, outperforming the state-of-the-art. We also show 3D skeleton prediction from varied and challenging 3D bodies. The code to infer a skeleton from a body shape is available for research at https://osso.is.tue.mpg.de/, and the dataset of paired outer surface (skin) and skeleton (bone) meshes is available as a Biobank Returned Dataset. This research has been conducted using the UK Biobank Resource.
Vertebrae segmentation, identification and localization using a graph optimization and a synergistic cycle
Oct 23, 2021
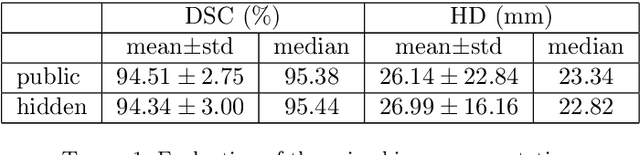
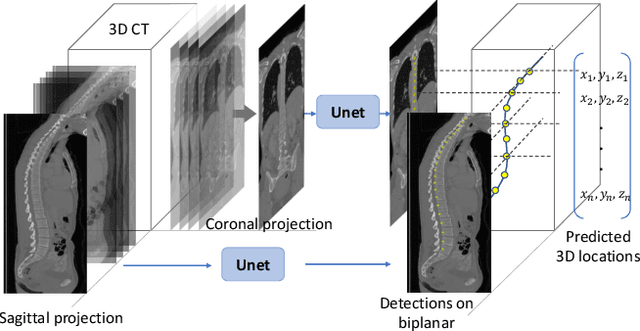
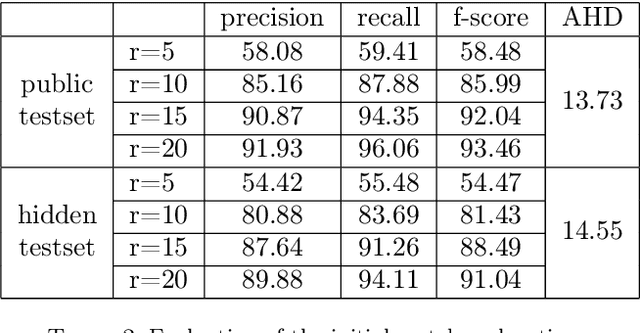
Abstract:This paper considers the segmentation, identification and localization of vertebrae in CT images. Although these three tasks are related, they face specific problems that add up when they are addressed together. For example neighboring vertebrae with similar shapes perturb the identification and vertebrae with complex or even pathological morphologies impact the segmentation. Consequently, the three tasks tend to be approached independently, e.g. labelling (localization and identification) or segmenting only, or, when treated globally, a sequential strategy is used. Sequential methods however are prone to accumulate errors as they are not able to recover from mistakes of the previous module. In this work, we propose to combine all three tasks and leverage their interdependence: locations ease the segmentation, the segmentations in turn improve the locations and they all contribute and benefit from the identification task. To this purpose we propose a virtuous cycle to enforce coherence between the three tasks. Within such a cycle, the tasks interoperate and are iterated until a global consistency criterion is satisfied. Our experiments validate this strategy with anatomically coherent results that outperform the state of the art on the VerSe20 challenge benchmark. Our code and model are openly available for research purposes at https://gitlab.inria.fr/spine/vertebrae_segmentation.
Dressing 3D Humans using a Conditional Mesh-VAE-GAN
Jul 31, 2019



Abstract:Three-dimensional human body models are widely used in the analysis of human pose and motion. Existing models, however, are learned from minimally-clothed humans and thus do not capture the complexity of dressed humans in common images and videos. To address this, we learn a generative 3D mesh model of clothing from 3D scans of people with varying pose. Going beyond previous work, our generative model is conditioned on different clothing types, giving the ability to dress different body shapes in a variety of clothing. To do so, we train a conditional Mesh-VAE-GAN on clothing displacements from a 3D SMPL body model. This generative clothing model enables us to sample various types of clothing, in novel poses, on top of SMPL. With a focus on clothing geometry, the model captures both global shape and local structure, effectively extending the SMPL model to add clothing. To our knowledge, this is the first conditional VAE-GAN that works on 3D meshes. For clothing specifically, it is the first such model that directly dresses 3D human body meshes and generalizes to different poses.
Learning and Tracking the 3D Body Shape of Freely Moving Infants from RGB-D sequences
Oct 17, 2018



Abstract:Statistical models of the human body surface are generally learned from thousands of high-quality 3D scans in predefined poses to cover the wide variety of human body shapes and articulations. Acquisition of such data requires expensive equipment, calibration procedures, and is limited to cooperative subjects who can understand and follow instructions, such as adults. We present a method for learning a statistical 3D Skinned Multi-Infant Linear body model (SMIL) from incomplete, low-quality RGB-D sequences of freely moving infants. Quantitative experiments show that SMIL faithfully represents the RGB-D data and properly factorizes the shape and pose of the infants. To demonstrate the applicability of SMIL, we fit the model to RGB-D sequences of freely moving infants and show, with a case study, that our method captures enough motion detail for General Movements Assessment (GMA), a method used in clinical practice for early detection of neurodevelopmental disorders in infants. SMIL provides a new tool for analyzing infant shape and movement and is a step towards an automated system for GMA.
Detailed, accurate, human shape estimation from clothed 3D scan sequences
Apr 19, 2017



Abstract:We address the problem of estimating human pose and body shape from 3D scans over time. Reliable estimation of 3D body shape is necessary for many applications including virtual try-on, health monitoring, and avatar creation for virtual reality. Scanning bodies in minimal clothing, however, presents a practical barrier to these applications. We address this problem by estimating body shape under clothing from a sequence of 3D scans. Previous methods that have exploited body models produce smooth shapes lacking personalized details. We contribute a new approach to recover a personalized shape of the person. The estimated shape deviates from a parametric model to fit the 3D scans. We demonstrate the method using high quality 4D data as well as sequences of visual hulls extracted from multi-view images. We also make available BUFF, a new 4D dataset that enables quantitative evaluation (http://buff.is.tue.mpg.de). Our method outperforms the state of the art in both pose estimation and shape estimation, qualitatively and quantitatively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge